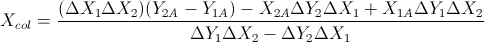The package ggsignif is very useful for quickly and easily indicating significant comparisons in ggplot graphs. However the comparisons call requires manual typing of each pair of values to be compared.
Eg.
library(ggplot2)
library(ggsignif)
data(iris)
ggplot(iris, aes(x=Species, y=Sepal.Length)) +
geom_boxplot() +
geom_signif(comparisons = list(c("versicolor", "virginica"),c('versicolor','setosa')),
map_signif_level=TRUE)
I'm wondering how this could be circumvented by referring to all the possible combinations at once? For example, expand.grid(x = levels(iris$Species), y = levels(iris$Species)), gives all the combinations
x y
1 setosa setosa
2 versicolor setosa
3 virginica setosa
4 setosa versicolor
5 versicolor versicolor
6 virginica versicolor
7 setosa virginica
8 versicolor virginica
9 virginica virginica
But how to have this accepted by geom_signif(comparisons=...?
Package info is available here https://cran.r-project.org/web/packages/ggsignif/index.html