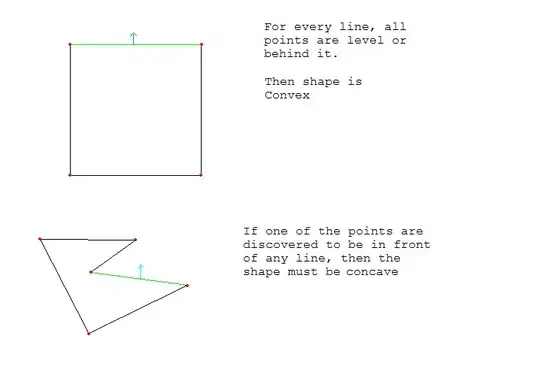
I'm stuck trying to fit a bipolar sigmoid curve - I'd like to have the following curve:
but I need it shifted and stretched. I have the following inputs:
x[0] = 8, x[48] = 2
So over 48 periods I need to drop from 8 to 2 using a bipolar sigmoid function to approximate a nice smooth dropoff. Any ideas how I could derive the curve that would fit those parameters?
Here's what I have so far, but I need to change the sigmoid function:
import math
def sigmoid(x):
return 1 / (1 + math.exp(-x))
plt.plot([sigmoid(float(z)) for z in range(1,48)])