I'm trying to implement the spiking neuron of the Izhikevich model. The formula for this type of neuron is really simple:
v[n+1] = 0.04*v[n]^2 + 5*v[n] + 140 - u[n] + I
u[n+1] = a*(b*v[n] - u[n])
where v is the membrane potential and u is a recovery variable.
If v gets above 30, it is reset to c and u is reset to u + d.
Given such a simple equation I wouldn't expect any problems. But while the graph should look like  , all I'm getting is this:
, all I'm getting is this: 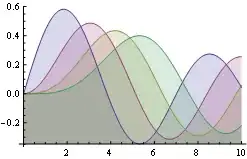
I'm completely at loss what I'm doing wrong exactly because there's so little to do wrong. I've looked for other implementations but the code I'm looking for is always hidden in a dll somewhere. However I'm pretty sure I'm doing exactly what the Matlab code of the author (2) is doing. Here is my full R code:
v = -70
u = 0
a = 0.02
b = 0.2
c = -65
d = 6
history <- c()
for (i in 1:100) {
if (v >= 30) {
v = c
u = u + d
}
v = 0.04*v^2 + 5*v + 140 - u + 0
u=a*(b*v-u);
history <- c(history, v)
}
plot(history, type = "l")
To anyone who's ever implemented an Izhikevich model, what am I missing?
usefull links: (1) http://www.opensourcebrain.org/projects/izhikevichmodel/wiki (2) http://www.izhikevich.org/publications/spikes.pdf
Answer
So it turns out I read the formula wrong. Apparently v' means new v = v + 0.04*v^2 + 5*v + 140 - u + I. My teachers would have written this as v' = 0.04*v^2 + 6*v + 140 - u + I. I'm very grateful for your help in pointing this out to me.