Currently I'm working with texts. My main goal is to calculate a measure of similarity between 30 000 texts. I'm following this tutorial:
Creating a document-term matrix:
In [1]: import numpy as np # a conventional alias
In [2]: from sklearn.feature_extraction.text import CountVectorizer
In [3]: filenames = ['data/austen-brontë/Austen_Emma.txt',
...: 'data/austen-brontë/Austen_Pride.txt',
...: 'data/austen-brontë/Austen_Sense.txt',
...: 'data/austen-brontë/CBronte_Jane.txt',
...: 'data/austen-brontë/CBronte_Professor.txt',
...: 'data/austen-brontë/CBronte_Villette.txt']
...:
In [4]: vectorizer = CountVectorizer(input='filename')
In [5]: dtm = vectorizer.fit_transform(filenames) # a sparse matrix
In [6]: vocab = vectorizer.get_feature_names() # a list
In [7]: type(dtm)
Out[7]: scipy.sparse.csr.csr_matrix
In [8]: dtm = dtm.toarray() # convert to a regular array
In [9]: vocab = np.array(vocab)
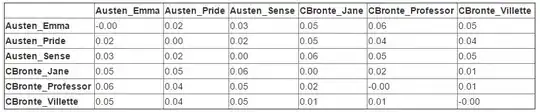
Comparing texts And we want to use a measure of distance that takes into consideration the length of the novels, we can calculate the cosine similarity.
In [24]: from sklearn.metrics.pairwise import cosine_similarity
In [25]: dist = 1 - cosine_similarity(dtm)
In [26]: np.round(dist, 2)
Out[26]:
array([[-0. , 0.02, 0.03, 0.05, 0.06, 0.05],
[ 0.02, 0. , 0.02, 0.05, 0.04, 0.04],
[ 0.03, 0.02, 0. , 0.06, 0.05, 0.05],
[ 0.05, 0.05, 0.06, 0. , 0.02, 0.01],
[ 0.06, 0.04, 0.05, 0.02, -0. , 0.01],
[ 0.05, 0.04, 0.05, 0.01, 0.01, -0. ]])
The final result:
As mentioned above, my goal is to calculate a measure of similarity between 30 000 texts. While implementing the above codes, it is taking too much time, in the end giving me a Memory Error. My question is --- Is there a better solution to calculate cosine similarity between huge number of texts? How do you cope with time and Memory Error Problem?