I implemented a multilayer perceptron with 1 hidden layer on MNIST dataset. The activation function in hidden layer is leaky(0.01) ReLu and output layer has a softmax activation function. The learning method is mini-batch SGD. The network structure is 784*30*10. The problem is I found the predictions the network made, for each input sample, are quite similar. That means the model would always like to think the image is some certain number. Thanks @Lemm Ras for pointing out the label-data mismatching problem in previous data_shuffle function and now fixed. But after some batch training, I found the predictions are still some kind of similar: 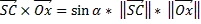 That's confusing.
That's confusing.
Another issue is the update value is too small comparing with original weight, in the MLP code, I add variable 'cc' and 'dd' to record the ratio between their weight_update and weight,
cc=W_OUTPUT_Update./W_OUTPUT;
dd=W_MLP_Update./W_MLP;
During debugging, the magnitude for cc is 10^-4(0.0001) and dd is also 10^-4. This might be the reason that the accuracy doesn't seems improved a lot.
After several days debugging. I have no idea why that happens and how to solve it, it made me stuck for one week. Can someone help me please? The screenshot is the value of A2 after softmax function.
[dimension, images, labels, labels_matrix, train_amount, test_labels_matrix, test_images, test_labels, test_amount] = load_mnist_data(); %initialize str
images=images(:,1:10000); % for debugging, get part of whole data set
labels=labels(1:10000,1);
labels_matrix=labels_matrix(:,1:10000);
test_images=test_images(:,1:500);
test_labels=test_labels(1:500,1);
train_amount=10000;
test_amount=500;
% initialize the structure
[ W_MAD, W_MLP, W_OUTPUT] = initialize_structure(dimension, train_amount, test_amount);
epoch=100;
correct_rate=zeros(1,epoch); %record testing accuracy
corr=zeros(1,epoch); %record training accuracy
lr=0.2;
lamda=0;
batch_size=50;
for i=1:epoch
sprintf('MLP in iteration %d over %d', i, epoch)
%shuffle data
[labels_shuffled labels_matrix_shuffled images_shuffled]=shuffle_data(labels, labels_matrix,images);
[ cor, W_MLP, W_OUTPUT ] = train_mlp_relu(lr, leaky, lamda, momentum_gamma, batch_size,W_MLP, W_OUTPUT, W_MAD, power, images_shuffled, train_amount, labels_shuffled, labels_matrix_shuffled);
corr(i)=cor/train_amount;
% test
correct_rate(i) = structure_test( W_MAD, W_MLP, W_OUTPUT, test_images, test_labels, test_amount );
end
% plot results
plot(1:epoch,correct_rate);
Here's the training MLP function, please ignore L2 regularization parameter lamda which is currently set as 0.
%MLP with batch size batch_size
cor=0;
%leaky=(1/batch_size);
leaky=0.001;
for i=1:train_amount/batch_size
batch_images=images(:,batch_size*(i-1)+1:batch_size*i);
batch_labels=labels_matrix(:,batch_size*(i-1)+1:batch_size*i);
%from MAD to MLP
V1=W_MLP'*batch_images;
V1(1,:)=1; %set bias unit as 1
V1_dirivative=ones(size(V1));
V1_dirivative(find(V1<0))=leaky;
A1=relu(V1,leaky); % A stands for activation
V2=W_OUTPUT'* A1;
A2=softmax(V2);
%write these scope control codes into functions.
%train error
[val idx]=max(A2);
idx=idx-1; %because index(idx) for matrix vaires from 1 to 10 while label varies from 0 to 9.
res=labels(batch_size*(i-1)+1:batch_size*i)-idx';
cor=cor+sum(res(:)==0);
%softmax loss, due to relu applied nodes that has
%contribution to activate neurons has gradient 1; while <0 nodes
%has no contribution
delta_softmax=-(1/batch_size)*(batch_labels-A2);
delta_output=W_OUTPUT*delta_softmax.*V1_dirivative;
%update
W_OUTPUT_Update=lr*(1/batch_size)*A1*delta_softmax'+lamda*W_OUTPUT;
cc=W_OUTPUT_Update./W_OUTPUT;
W_MLP_Update=lr*(1/batch_size)*batch_images*delta_output'+lamda*W_MLP;
dd=W_MLP_Update./W_MLP;
k=mean(A2,2);
W_OUTPUT=W_OUTPUT-W_OUTPUT_Update;
W_MLP=W_MLP-W_MLP_Update;
end
end
Here is the softmax function:
function [ val ] = softmax( val )
val=exp(val);
val=val./repmat(sum(val),10,1);
end
The labels_matrix is the aimed output matrix for A2 and created as:
labels_matrix=full(sparse(labels+1,1:train_amount,1));
test_labels_matrix=full(sparse(test_labels+1,1:test_amount,1));
And Relu:
function [ val ] = relu( val,leaky )
val(find(val<0))=leaky*val(find(val<0));
end
Data shuffle
%this version is wrong, due to it only shuffles label and data without doing the same shuffling on the 'labels_matrix' which is used to calculate MLP's delta in output layer. It destroyed the link between data and label.
% function [ label, data ] = shuffle_data( label, data )
% [row column]=size(data);
% array=randperm(column);
% data=data(:,array);
% label=label(array);
% %if shuffle respect to row then use the code below
% %data=data(randperm(row),:);
% end
function [ label, label_matrix, data ] = shuffle_data( label, label_matrix, data )
[row column]=size(data);
array=randperm(column);
data=data(:,array);
label=label(array);
label_matrix=label_matrix(:, array);
%if shuffle respect to row then use the code below
%data=data(randperm(row),:);
end
Data loading:
function [ dimension, images, labels, labels_matrix, train_amount, test_labels_matrix, test_images, test_labels, test_amount] = load_mnist_data()
%%load training and testing data, labels
data_location='C:\Users\yz39g15\Documents\MATLAB\common\mnist test\for the report/modify/train-images.idx3-ubyte';
label_location='C:\Users\yz39g15\Documents\MATLAB\common\mnist test\for the report/modify/train-labels.idx1-ubyte';
test_data_location='C:\Users\yz39g15\Documents\MATLAB\common\mnist test\for the report/modify/t10k-images.idx3-ubyte';
test_label_location='C:\Users\yz39g15\Documents\MATLAB\common\mnist test\for the report/modify/t10k-labels.idx1-ubyte';
images = loadMNISTImages(data_location);
labels = loadMNISTLabels(label_location);
test_images=loadMNISTImages(test_data_location);
test_labels=loadMNISTLabels(test_label_location);
%%data centralization
[dimension train_amount]=size(images);
[dimension test_amount]=size(test_images);
%%complete normalization
%%transform labels from index to matrix in order to apply square loss function in output layer
labels_matrix=full(sparse(labels+1,1:train_amount,1));
test_labels_matrix=full(sparse(test_labels+1,1:test_amount,1));
end