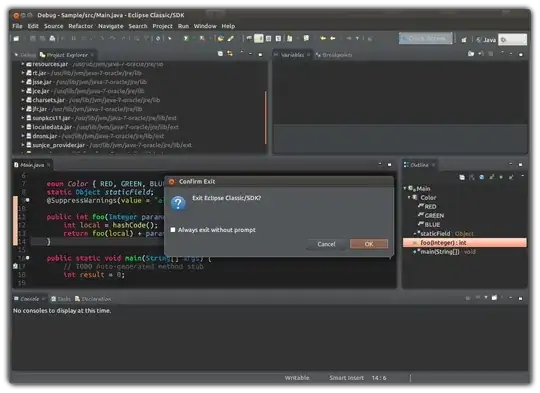
I'm trying to plot my data so that it looks something like this (taken from this blog):

My data looks like this:
head(Diet)
Group Category studies_n studies_pc
1 algae algae 61 38.4
2 algae biofilm 4 2.5
3 algae diatoms 8 5.0
4 algae fil alg 18 11.3
5 algae phytoplankton 2 1.3
6 insect Coleoptera 59 37.1
"Category" is actually a sub-category of "Group", and each "Group" contains distinct "Categories". For example, the algae Group only contains those five different kinds of algae listed in "Category", and the insect group doesn't include any algae categories.
I want the Group to be plotted in a specific order and the number of studies ("studies_n") to be plotted in rank order:
Diet$Group <- factor(Diet$Group, levels = c("algae", "detritus", "mollusc", "crustacean", "insect", "fish", "other"))
Diet$Category <- factor(Diet$Category, levels=Diet[order(Diet$studies_n), "Category"])
With this established, I have tried two different ways to plot the data:
METHOD A.
pDiet <- ggplot(Diet, aes(x=Category, weight=studies_n)) +
geom_bar() +
facet_grid(.~Group, scales="free_x", space="free") +
ylab("number of studies") +
theme(axis.text.x=element_text(angle=90))
plot(pDiet)
This is ok, but I would rather the bars be horizontal than vertical (to make it easier to read the labels). Also, the text on the x-axis is not aligned.
METHOD B.
pDiet <- ggplot(Diet, aes(x=Category, y=studies_n)) +
geom_bar(stat="identity") +
coord_flip() +
theme(axis.title.y=element_blank()) +
facet_grid(Group~., scales="free", space="free") +
theme(strip.text.y = element_text(angle=0)) +
ylab("number of studies")
plot(pDiet)
This is a better layout, but all of the Categories are plotted for each Group and that's just ridiculous.
Help?