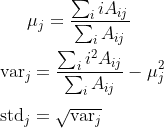You want to take the mean, variance and standard deviation of the vector [1, 2, 3, ..., n] — where n is the dimension of the input matrix A along the axis of interest —, with weights given by the matrix A itself.
For concreteness, say you want to consider these center-of-mass statistics along the vertical axis (axis=0) — this is what corresponds to the formulas you wrote. For a fixed column j, you would do
n = A.shape[0]
r = np.arange(1, n+1)
mu = np.average(r, weights=A[:,j])
var = np.average(r**2, weights=A[:,j]) - mu**2
std = np.sqrt(var)
In order to put all of the computations for the different columns together, you have to stack together a bunch of copies of r (one per column) to form a matrix (that I have called R in the code below). With a bit of care, you can make things work for both axis=0 and axis=1.
import numpy as np
def com_stats(A, axis=0):
A = A.astype(float) # if you are worried about int vs. float
n = A.shape[axis]
m = A.shape[(axis-1)%2]
r = np.arange(1, n+1)
R = np.vstack([r] * m)
if axis == 0:
R = R.T
mu = np.average(R, axis=axis, weights=A)
var = np.average(R**2, axis=axis, weights=A) - mu**2
std = np.sqrt(var)
return mu, var, std
For example,
A = np.array([[1, 1, 0], [1, 2, 1], [1, 1, 1]])
print(A)
# [[1 1 0]
# [1 2 1]
# [1 1 1]]
print(com_stats(A))
# (array([ 2. , 2. , 2.5]), # centre-of-mass mean by column
# array([ 0.66666667, 0.5 , 0.25 ]), # centre-of-mass variance by column
# array([ 0.81649658, 0.70710678, 0.5 ])) # centre-of-mass std by column
EDIT:
One can avoid creating in-memory copies of r to build R by using numpy.lib.stride_tricks: swap the line
R = np.vstack([r] * m)
above with
from numpy.lib.stride_tricks import as_strided
R = as_strided(r, strides=(0, r.itemsize), shape=(m, n))
The resulting R is a (strided) ndarray whose underlying array is the same as r's — absolutely no copying of any values occurs.
from numpy.lib.stride_tricks import as_strided
FMT = '''\
Shape: {}
Strides: {}
Position in memory: {}
Size in memory (bytes): {}
'''
def find_base_nbytes(obj):
if obj.base is not None:
return find_base_nbytes(obj.base)
return obj.nbytes
def stats(obj):
return FMT.format(obj.shape,
obj.strides,
obj.__array_interface__['data'][0],
find_base_nbytes(obj))
n=10
m=1000
r = np.arange(1, n+1)
R = np.vstack([r] * m)
S = as_strided(r, strides=(0, r.itemsize), shape=(m, n))
print(stats(r))
print(stats(R))
print(stats(S))
Output:
Shape: (10,)
Strides: (8,)
Position in memory: 4299744576
Size in memory (bytes): 80
Shape: (1000, 10)
Strides: (80, 8)
Position in memory: 4304464384
Size in memory (bytes): 80000
Shape: (1000, 10)
Strides: (0, 8)
Position in memory: 4299744576
Size in memory (bytes): 80
Credit to this SO answer and this one for explanations on how to get the memory address and size of the underlying array of a strided ndarray.