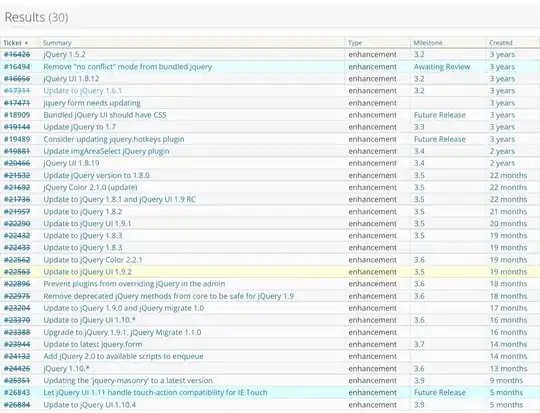
R:
df looks like this (x = a, y = b, group = c):
a b c
-------------------
-2.1 1203 5
1.4 1103 10
-2.1 1203 5
.. .. ..
I created a scatterplot with around 10 linear regression lines (ggplot2/geom_smooth) from this df, for each group in c(e.g. 5, 10). These have all different slopes and y-intersects
Is there any way I can approximate the function for the family of curves for these linear regression lines in R and display it with custom parameters in a new plot(ggplot2)?
edit:
Here is the dput(df): (will remove it again later)
structure(list(X = 1:102, a = c("0", "0", "0", "0", "0", "219.399.914.550.781",
"987", "0", "0", "0", "0", "1320", "0", "0", "0", "0", "0", "0",
"0", "0", "0", "0", "0", "0", "0", "0", "0", "144.595.013.427.734",
"176.470.013.427.734", "167.919.995.117.188", "125.239.242.553.711",
"247.582.397.460.938", "173.550.708.007.812", "149.010.693.359.375",
"908", "638.5", "81.173.999.023.438", "1472", "120.632.000.732.422",
"2028", "154.019.999.694.824", "785.5", "118.188.000.488.281",
"149.930.010.986.328", "-712", "-2100", "1628", "925", "1161",
"516", "64.840.002.441.406", "426.5", "106.810.998.535.156",
"92.175.994.873.047", "648.5", "125.379.998.779.297", "1120",
"821", "795", "129.179.998.779.297", "137.460.000.610.352", "127.231.660.461.426",
"148.579.998.779.297", "115.440.997.314.453", "4.482.857.905.469",
"1163", "97.440.002.441.406", "130.298.852.539.062", "195.956.491.088.867",
"504.998.779.296.989", "1043", "228.998.406.982.422", "128.374.566.650.391",
"153.219.995.117.188", "111.604.742.431.641", "108.100.006.103.516",
"1364.5", "102.669.999.694.824", "141.820.001.220.703", "83.124.743.652.344",
"93.209.649.658.203", "149.629.656.982.422", "150.215.002.441.406",
"161.379.998.779.297", "41.129.998.779.297", "91.320.001.220.703",
"83.047.998.046.875", "1144.5", "104.020.001.220.703", "171.528.002.929.688",
"1519", "123.510.003.662.109", "106.240.002.441.406", "145.934.997.558.594",
"177.939.999.389.648", "195.360.003.662.109", "164.140.002.441.406",
"113.640.002.441.406", "146.676.000.976.562", "1.769.916.015.625",
"53.389.654.541.016", "685.018.981.933.594"), c = c(88L, 88L,
88L, 88L, NA, 88L, 88L, 88L, NA, NA, NA, 86L, 86L, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 90L, 88L, 88L, 88L,
88L, 88L, 88L, 88L, 88L, 88L, 88L, 88L, 86L, 88L, 88L, 88L, 84L,
84L, 84L, 84L, 86L, 84L, 84L, 84L, 84L, 84L, 84L, 84L, 84L, 84L,
82L, 82L, 84L, 82L, 82L, 84L, 82L, 82L, 82L, 82L, 82L, 82L, 82L,
82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 80L, 80L,
80L, 80L, 80L, 82L, 84L, 82L, 82L, 80L, 80L, 80L, 80L, 80L, 80L,
80L, 80L, 80L, 80L, 80L, 80L), c_null = c(88L, 88L, 88L, 88L,
0L, 88L, 88L, 88L, 0L, 0L, 0L, 86L, 86L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 90L, 88L, 88L, 88L, 88L,
88L, 88L, 88L, 88L, 88L, 88L, 88L, 86L, 88L, 88L, 88L, 84L, 84L,
84L, 84L, 86L, 84L, 84L, 84L, 84L, 84L, 84L, 84L, 84L, 84L, 82L,
82L, 84L, 82L, 82L, 84L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L,
82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 82L, 80L, 80L, 80L,
80L, 80L, 82L, 84L, 82L, 82L, 80L, 80L, 80L, 80L, 80L, 80L, 80L,
80L, 80L, 80L, 80L, 80L), c_orig = c("88.000.096.643.065", "88.000.096.643.065",
"88.000.096.643.065", "0", "874.979.654.044.919", "873.618.932.305.081",
"869.990.179.502.541", "0", "0", "0", "861.825.503", "861.825.503",
"0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0",
"0", "899.000.015.258.789", "87.5", "880.999.984.741.211", "88",
"879.000.015.258.789", "87", "869.000.015.258.789", "87", "868.000.030.517.578",
"878.000.030.517.578", "876.999.969.482.422", "865.999.984.741.211",
"861.999.969.482.422", "870.999.984.741.211", "869.000.015.258.789",
"865.999.984.741.211", "871.999.969.482.422", "841.999.969.482.422",
"84.5", "840.999.984.741.211", "845.999.984.741.211", "843.000.030.517.578",
"841.999.969.482.422", "83", "834.000.015.258.789", "83", "825.999.984.741.211",
"834.000.015.258.789", "831.999.969.482.422", "826.999.969.482.422",
"823.000.030.517.578", "821.999.969.482.422", "825.999.984.741.211",
"821.999.969.482.422", "82", "825.999.984.741.211", "816.999.969.482.422",
"814.000.015.258.789", "81", "819.000.015.258.789", "816.999.969.482.422",
"81.5", "821.999.969.482.422", "811.999.969.482.422", "814.000.015.258.789",
"813.000.030.517.578", "808.000.030.517.578", "815.999.984.741.211",
"818.000.030.517.578", "814.000.015.258.789", "814.000.015.258.789",
"809.000.015.258.789", "809.000.015.258.789", "805.999.984.741.211",
"801.999.969.482.422", "796.999.969.482.422", "801.999.969.482.422",
"803.000.030.517.578", "804.000.015.258.789", "811.999.969.482.422",
"825.999.984.741.211", "82.5", "819.000.015.258.789", "804.000.015.258.789",
"795.999.984.741.211", "804.000.015.258.789", "80", "801.999.969.482.422",
"798.000.030.517.578", "80", "80", "795.999.984.741.211", "800.999.984.741.211",
"799.000.015.258.789", "791.999.969.482.422", "791.999.969.482.422"
), b = c("0", "0", "0", NA, NA, "-0.136072173983791", "-0.362875280254002",
NA, NA, NA, NA, "0", NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, "-240.000.152.587.891", "0.599998474121094",
"-0.0999984741210938", "-0.0999984741210938", "-0.900001525878906",
"-0.0999984741210938", "0.0999984741210938", "-0.199996948242202",
"1", "-0.100006103515597", "-109.999.847.412.111", "-0.400001525878892",
"0.900001525878892", "-0.199996948242188", "-0.300003051757812",
"0.599998474121108", "0.5", "0.300003051757798", "-0.400001525878906",
"0.5", "-0.299995422363295", "-0.100006103515597", "-11.999.969.482.422",
"0.400001525878906", "-0.400001525878906", "-0.400001525878906",
"0.800003051757812", "-0.200004577636705", "-0.5", "-0.399993896484403",
"-0.100006103515597", "0.400001525878892", "-0.400001525878892",
"-0.199996948242202", "0.599998474121094", "-0.900001525878892",
"-0.299995422363295", "-0.400001525878906", "0.900001525878906",
"-0.200004577636705", "-0.199996948242202", "0.699996948242202",
"-1", "0.200004577636705", "-0.099998474121108", "-0.5", "0.799995422363295",
"0.200004577636705", "-0.400001525878892", "0", "-0.5", "0",
"-0.300003051757812", "-0.400001525878892", "-0.5", "0.5", "0.100006103515597",
"0.099998474121108", "0.799995422363295", "140.000.152.587.889",
"-0.0999984741210938", "-0.599998474121094", "-1.5", "-0.800003051757812",
"0.800003051757812", "-0.400001525878906", "0.199996948242202",
"-0.399993896484403", "0.199996948242202", "0", "-0.400001525878906",
"0.5", "-0.199996948242188", "-0.700004577636705", NA), b_null = c("0",
"0", "0", "0", "0", "-0.136072173983791", "-0.362875280254002",
"0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0",
"0", "0", "0", "0", "0", "0", "0", "-240.000.152.587.891", "0.599998474121094",
"-0.0999984741210938", "-0.0999984741210938", "-0.900001525878906",
"-0.0999984741210938", "0.0999984741210938", "-0.199996948242202",
"1", "-0.100006103515597", "-109.999.847.412.111", "-0.400001525878892",
"0.900001525878892", "-0.199996948242188", "-0.300003051757812",
"0.599998474121108", "0.5", "0.300003051757798", "-0.400001525878906",
"0.5", "-0.299995422363295", "-0.100006103515597", "-11.999.969.482.422",
"0.400001525878906", "-0.400001525878906", "-0.400001525878906",
"0.800003051757812", "-0.200004577636705", "-0.5", "-0.399993896484403",
"-0.100006103515597", "0.400001525878892", "-0.400001525878892",
"-0.199996948242202", "0.599998474121094", "-0.900001525878892",
"-0.299995422363295", "-0.400001525878906", "0.900001525878906",
"-0.200004577636705", "-0.199996948242202", "0.699996948242202",
"-1", "0.200004577636705", "-0.099998474121108", "-0.5", "0.799995422363295",
"0.200004577636705", "-0.400001525878892", "0", "-0.5", "0",
"-0.300003051757812", "-0.400001525878892", "-0.5", "0.5", "0.100006103515597",
"0.099998474121108", "0.799995422363295", "140.000.152.587.889",
"-0.0999984741210938", "-0.599998474121094", "-1.5", "-0.800003051757812",
"0.800003051757812", "-0.400001525878906", "0.199996948242202",
"-0.399993896484403", "0.199996948242202", "0", "-0.400001525878906",
"0.5", "-0.199996948242188", "-0.700004577636705", "0")), .Names = c("X",
"a", "c", "c_null", "c_orig", "b", "b_null"), class = "data.frame", row.names = c(NA,
-102L))
And I have just plotted it with ggplot2:
ggplot(aes(x = a, y = b, group = c), data = Health, na.rm = TRUE) +
geom_point(aes(color = c, size = factor(c)), alpha=0.3) +
scale_color_distiller(palette="YlGnBu", na.value="white") +
geom_smooth(method = "lm", aes(group = factor(c), color = c), se = F)
And now I want to approximate the family of curves for all the geom_smooth lines to predict in another plot different scenarios with other parameters!