I tried to reproduce ROC curve from the plot.roc function from pRoc package with ggplot2.
library(mlbench)
library(caret)
data(Sonar)
set.seed(998)
fitControl <- trainControl(method = "repeatedcv",
number = 10,
repeats = 10,
## Estimate class probabilities
classProbs = TRUE,
## Evaluate performance using
## the following function
summaryFunction = twoClassSummary)
gbmGrid <- expand.grid(interaction.depth = c(1, 5, 9),
n.trees = (1:30)*50,
shrinkage = 0.1,
n.minobsinnode = 20)
inTraining <- createDataPartition(Sonar$Class, p = .75, list = FALSE)
training <- Sonar[ inTraining,]
testing <- Sonar[-inTraining,]
set.seed(825)
gbmFit <- train(Class ~ ., data = training,
method = "gbm",
trControl = fitControl,
verbose = FALSE,
tuneGrid = gbmGrid,
## Specify which metric to optimize
metric = "ROC")
gbmFit
probs = predict(gbmFit, newdata = testing, type = "prob")
roc = roc(predictor = probs$M,
response = testing$Class,
levels = c('M','R'),
percent = TRUE)
plot.roc(roc, print.auc = TRUE, col='red')
df = data.frame(Specificity=roc$specificities, Sensitivity=roc$sensitivities)
ggplot(data = df, aes(x = Specificity, y = Sensitivity))+
geom_step(color='red', size=2, direction = "hv")+
scale_x_reverse()+
geom_abline(intercept = 100, slope = 1, color='grey')+
annotate("text", x = 30, y = 20, label = paste0('AUC: ', round(roc$auc,1), '%'), size = 8)+
ylab('Sensitivity (%)')+
xlab('Specificity (%)')
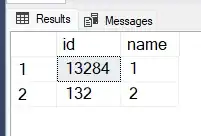
scale_x_reverse() seems to be the problem, is there any other way to reverse the X axis or to correct that plot?