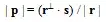I have the following distance matrix:
delta =
[[ 0. 0.71370845 0.80903791 0.82955157 0.56964983 0. 0. ]
[ 0.71370845 0. 0.99583115 1. 0.79563006 0.71370845
0.71370845]
[ 0.80903791 0.99583115 0. 0.90029133 0.81180111 0.80903791
0.80903791]
[ 0.82955157 1. 0.90029133 0. 0.97468433 0.82955157
0.82955157]
[ 0.56964983 0.79563006 0.81180111 0.97468433 0. 0.56964983
0.56964983]
[ 0. 0.71370845 0.80903791 0.82955157 0.56964983 0. 0. ]
[ 0. 0.71370845 0.80903791 0.82955157 0.56964983 0. 0. ]]
And I'm trying to use the networkx library to represent it as a graph. This is my code:
import networkx as nx
G = nx.from_numpy_matrix(delta)
pos = nx.random_layout(G)
plt.figure(figsize=(7, 7))
for k, p in pos.iteritems():
plt.scatter(p[0], p[1], marker='o', c=colors[k], s=50, edgecolor='None')
lgd = plt.legend(markers, labels, numpoints=1, bbox_to_anchor=(1.17, 0.5))
plt.tight_layout()
plt.axis('equal')
pt.show()
However, what I see is not what I expect. For instance, consider this output:
From delta, node 1 is at the same point as node 6 and 7, and far from node 4. I don't see the in the output plot. Besides, overtime I run it, it results in another output. This is expected, but the distance seem to not be respected. In the following plot, for example, distances between 1 to 6,7 and 4 changed.
I can't understand why.