This is my first time of doing visualization from hierarchical data in dictionary format with Python. Last part of the data looks like this:
d = {^2820: [^391, ^1024], ^2821: [^759, 'w', ^118, ^51], ^2822: [^291, 'o'], ^2823: [^25, ^64], ^2824: [^177, ^2459], ^2825: [^338, ^1946], ^2826: [^186, ^1511], ^2827: [^162, 'i']}
So I have indices on lists referring back to the keys (index) of the dictionary. I suppose this could be used as a base structure for the visualization, please correct me if I'm wrong. Characters on the data are "end nodes/leaves" which doesn't refer back to any index.
I have found NetworkX which possibly could be used for visualization, but I have no idea where to start with it and my data. I was hoping it would be something as simple as:
import networkx as nx
import matplotlib.pyplot as plt
d = {^2820: [^391, ^1024], ^2821: [^759, 'w', ^118, ^51], ^2822: [^291, 'o'], ^2823: [^25, ^64], ^2824: [^177, ^2459], ^2825: [^338, ^1946], ^2826: [^186, ^1511], ^2827: [^162, 'i']}
G = nx.Graph(d)
nx.draw(G)
plt.show()
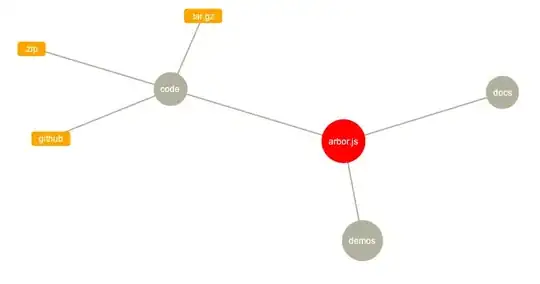
I'm looking for hierarchical dendrogram or some sort of clustering as an output. Sorry at this point I'm not totally sure what would be the best visualization, maybe similar to this:
UPDATE
Using NetworkX actually was very simple. I'm providing other simple sample data and looking for an answer if it can be visualized by dendrogram also instead of wired network graph?
# original sequence: a,b,c,d,b,c,a,b,c,d,b,c
d = {^1: ['b', 'c'], ^2: ['a', ^1, 'd', ^1], 'S': [^2, ^2]}
G = nx.Graph(d)
nx.draw_spring(G, node_size=300, with_labels=True)
As we can see, graph show plain relations, but not hierarchy and order of the data what I'm willing to do. DiGraph gives more details, but it is still not possible to construct original sequence from it:
For dendrogram apparently weight and end nodes needs to be calculated as pointed out on the first answer. For that approach data structure could be something like this:
d = {'a': [], 'b': [], 'c': [], 'd': [], ^1: ['b', 'c'], ^2: ['a', ^1, 'd', ^1], 'S': [^2, ^2]}