I built a wrapped bivariate gaussian distribution in Python using the equation given here: http://www.aos.wisc.edu/~dvimont/aos575/Handouts/bivariate_notes.pdf However, I don't understand why my distribution fails to sum to 1 despite having incorporated a normalization constant.
For a U x U lattice,
import numpy as np
from math import *
U = 60
m = np.arange(U)
i = m.reshape(U,1)
j = m.reshape(1,U)
sigma = 0.1
ii = np.minimum(i, U-i)
jj = np.minimum(j, U-j)
norm_constant = 1/(2*pi*sigma**2)
xmu = (ii-0)/sigma; ymu = (jj-0)/sigma
rhs = np.exp(-.5 * (xmu**2 + ymu**2))
ker = norm_constant * rhs
>> ker.sum() # area of each grid is 1
15.915494309189533
I'm certain there's fundamentally missing in the way I'm thinking about this and suspect that some sort of additional normalization is needed, although I can't reason my way around it.
UPDATE:
Thanks to others' insightful suggestions, I rewrote my code to apply L1 normalization to kernel. However, it appears that, in the context of 2D convolution via FFt, keeping the range as [0, U] is able to still return a convincing result:
U = 100
Ukern = np.copy(U)
#Ukern = 15
m = np.arange(U)
i = m.reshape(U,1)
j = m.reshape(1,U)
sigma = 2.
ii = np.minimum(i, Ukern-i)
jj = np.minimum(j, Ukern-j)
xmu = (ii-0)/sigma; ymu = (jj-0)/sigma
ker = np.exp(-.5 * (xmu**2 + ymu**2))
ker /= np.abs(ker).sum()
''' Point Density '''
ido = np.random.randint(U, size=(10,2)).astype(np.int)
og = np.zeros((U,U))
np.add.at(og, (ido[:,0], ido[:,1]), 1)
''' Convolution via FFT and inverse-FFT '''
v1 = np.fft.fft2(ker)
v2 = np.fft.fft2(og)
v0 = np.fft.ifft2(v2*v1)
dd = np.abs(v0)
plt.plot(ido[:,1], ido[:,0], 'ko', alpha=.3)
plt.imshow(dd, origin='origin')
plt.show()
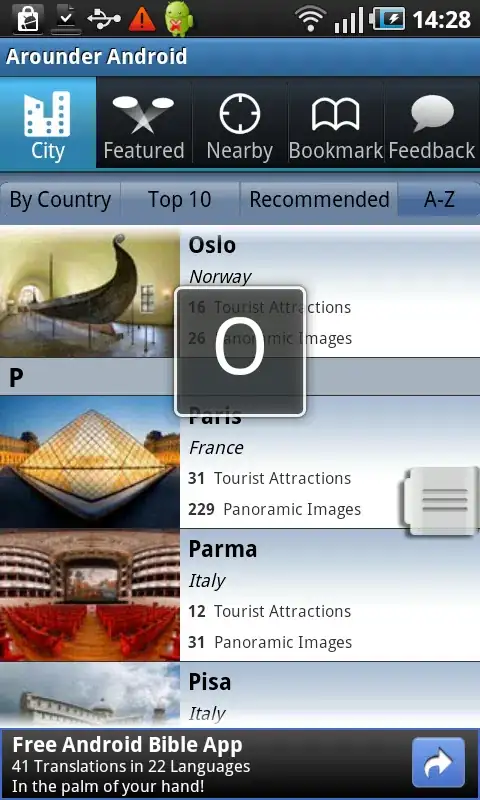 On the other hand, sizing the kernel using the commented-out line gives this incorrect plot:
On the other hand, sizing the kernel using the commented-out line gives this incorrect plot: