How about this:
Cook-up some data (i was hoping you would cook it for me :) )
library(igraph)
set.seed(666)
# herbivore-plant
m_hp <- matrix( rbinom(12, 10, p=0.2), 4, 3)
dimnames(m_hp) <- list(
consuming=paste0("h", seq(1, nrow(m_hp))),
consumed=paste0("p", seq(1, ncol(m_hp)))
)
# carnivore-herbivore
m_ch <- matrix( rbinom(20, 10, p=0.2), 5, 4)
dimnames(m_ch) <- list(
consuming=paste0("c", seq(1, nrow(m_ch))),
consumed=paste0("h", seq(1, ncol(m_ch)))
)
... so it looks like yours (I presume):
m_hp
## consumed
## consuming p1 p2 p3
## h1 3 1 0
## h2 1 3 1
## h3 5 5 3
## h4 1 2 0
m_ch
## consumed
## consuming h1 h2 h3 h4
## c1 0 4 0 2
## c2 1 2 1 2
## c3 1 2 3 3
## c4 3 5 1 2
## c5 0 2 0 2
Now turn them into igraph objects via edgelists
el_hp <- as.data.frame(as.table(m_hp), stringsAsFactors = FALSE)
el_ch <- as.data.frame(as.table(m_ch), stringsAsFactors = FALSE)
el <- rbind(el_hp, el_ch)
g <- graph.data.frame( el[el$Freq != 0 , ] )
V(g)$type <- substr(V(g)$name, 1, 1)
Adjacency matrix of joint network:
get.adjacency(g, sparse=FALSE, attr="Freq")
## h1 h2 h3 h4 c2 c3 c4 c1 c5 p1 p2 p3
## h1 0 0 0 0 0 0 0 0 0 3 1 0
## h2 0 0 0 0 0 0 0 0 0 1 3 1
## h3 0 0 0 0 0 0 0 0 0 5 5 3
## h4 0 0 0 0 0 0 0 0 0 1 2 0
## c2 1 2 1 2 0 0 0 0 0 0 0 0
## c3 1 2 3 3 0 0 0 0 0 0 0 0
## c4 3 5 1 2 0 0 0 0 0 0 0 0
## c1 0 4 0 2 0 0 0 0 0 0 0 0
## c5 0 2 0 2 0 0 0 0 0 0 0 0
## p1 0 0 0 0 0 0 0 0 0 0 0 0
## p2 0 0 0 0 0 0 0 0 0 0 0 0
## p3 0 0 0 0 0 0 0 0 0 0 0 0
Graphically
t <- match(V(g)$type, c("p", "h", "c") )
plot(g, vertex.color=t )
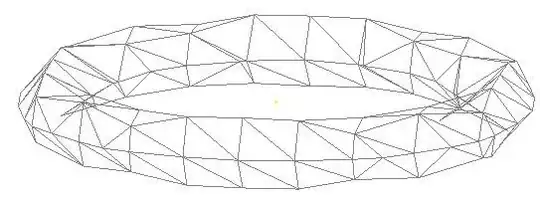
or even
l <- layout_with_fr(g, miny=t, maxy=t )
plot(g, vertex.color=t, layout=l, edge.width=E(g)$Freq)