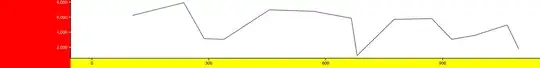
I am having issues limiting the regression lines in my "visreg" plots to the range of the data for each plot. They currently plot to the extent of the overall data frame and when I change the individual limits, it still plots to the overall data frame visually but not the correct data points. My sample codes are below:
CODrysub
Humid Wet.sub.himid Dry.sub.himid Semi.arid Arid Hyper.arid
Nov 2004 2.20 2.22 2.16 2.03 1.79 1.68
Dec 2004 2.75 2.72 2.52 2.32 2.08 1.93
Jan 2005 2.98 2.92 2.68 2.43 2.18 2.03
Feb 2005 2.81 2.80 2.62 2.37 2.14 2.04
Mar 2005 2.43 2.44 2.33 2.16 1.99 1.96
Apr 2005 2.30 2.25 2.08 2.00 1.90 1.91
SMDrysub
Humid Wet.sub.himid Dry.sub.himid Semi.arid Arid Hyper.arid
Nov 2004 0.26 0.21 0.15 0.08 0.05 0.07
Dec 2004 0.22 0.16 0.10 0.07 0.06 0.08
Jan 2005 0.19 0.12 0.08 0.07 0.07 0.09
Feb 2005 0.18 0.11 0.08 0.07 0.08 0.10
Mar 2005 0.18 0.13 0.09 0.07 0.07 0.10
Apr 2005 0.19 0.15 0.10 0.07 0.06 0.08
library(visreg)
Humida<-cbind(CODrysub[,1],SMDrysub[,1])
Humid2a<-as.data.frame(Humida)
colnames(Humid2a)<-c("CO","SM")
fit1a<-lm(CO~SM,data=Humid2a)
WSHa<-cbind(CODrysub[,2],SMDrysub[,2])
WSH2a<-as.data.frame(WSHa)
colnames(WSH2a)<-c("CO","SM")
fit2a<-lm(CO~SM,data=WSH2a)
##To plot
## Overall plot of an empty frame with dataframe labels
plot(0,xlim=c(0.05,0.31),ylim=c(1.5,3.4),cex.main=1, main= "(a) Scatterplots without soil moisture (Dry season)",xlab=NA,ylab=NA)
par(new=T)
visreg(fit1a,xlab="SM",ylab="CO",points=list(cex=1.2, pch=20,col="deepskyblue4") ,alpha=0.8,line=list(col="deepskyblue4",lwd=2),fill=list(col=adjustcolor("deepskyblue4", alpha.f = 0.09)), xlim=c(0.05,0.31),ylim=c(1.5,3.4),xaxt="n",yaxt="n")
par(new=T)
visreg(fit2a,xlab=NA,ylab=NA,points=list(cex=1.2, pch=20,col="dodgerblue1") ,line=list(col="dodgerblue1",lwd=2),fill=list(col=adjustcolor("dodgerblue1", alpha.f = 0.09)) ,xlim=c(0.05,0.31),ylim=c(1.5,3.4),alpha=0.8,xaxt="n",yaxt="n")
When I plot it this way, the regression lines go to infinity especially for data in the left corner of my plot. I have also tried defining the individual data limits but it plots my points to the extent of the data frame which is wrong. How do I limit the regression lines to the data extent, please?