In sklearn, GridSearchCV can take a pipeline as a parameter to find the best estimator through cross validation. However, the usual cross validation is like this:
to cross validate a time series data, the training and testing data are often splitted like this: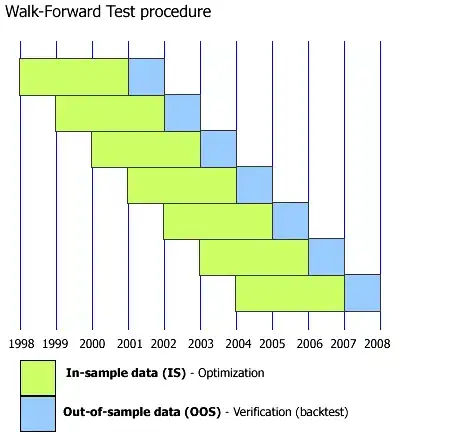
That is to say, the testing data should be always ahead of training data.
My thought is:
Write my own version class of k-fold and passing it to GridSearchCV so I can enjoy the convenience of pipeline. The problem is that it seems difficult to let GridSearchCV to use an specified indices of training and testing data.
Write a new class GridSearchWalkForwardTest which is similar to GridSearchCV, I am studying the source code grid_search.py and find it is a little complicated.
Any suggestion is welcome.