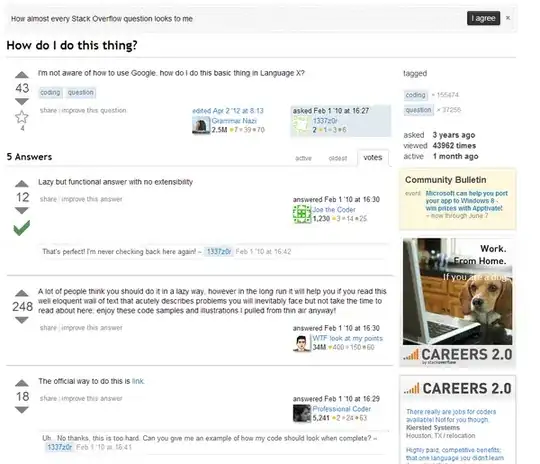
I had following
RMSE Model fold
1 0.4164260 lm 1
2 0.5412474 lm 2
3 0.3496156 lm 3
4 0.4431751 lm 4
5 0.4082882 lm 5
6 0.4398985 lm 6
7 0.4506528 lm 7
8 0.4236024 lm 8
9 0.4346417 lm 9
10 0.4982427 lm 10
11 0.3993842 MARS 1
12 0.5684007 MARS 2
13 0.3487462 MARS 3
14 0.4174607 MARS 4
15 0.3930033 MARS 5
16 0.4456201 MARS 6
17 0.4373339 MARS 7
18 0.4110089 MARS 8
19 0.4249311 MARS 9
20 0.4295155 MARS 10
I am plotting as follows:
ggplot(df, aes(x=factor(fold), y=RMSE, group=Model, colour=Model)) + geom_line()
I want to do mean line for both models. How can I achieve that?