I have a rather large tree like structure / dendrogram like / web (think pedigree) and I want to create a list of singularly connected leafs / nodes.
In genealogy something similar I believe is called a "Spitzenahnen" (German) / but it I believe is specific to 'no known parents', not necessarily no descendants. So basically dead ends in the structure, not just top or bottom is what I am looking to find.
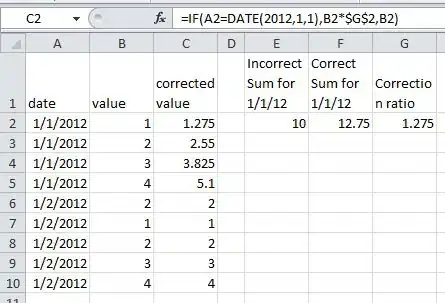
I saw the post on creating a edge list from a Matrix as well as how to access the attributes of a dendrogram in R but not sure how to apply it to get the specific results I am looking to obtain.
I have thousands of nodes with multiple starting and end points. I want to create a list of nodes/leafs where there is only one connected node that attaches it to the tree. So if there are two or more connections to the node (some have up to two dozen at most), I do not want to see it in my list.
Using a marked up graphic from "Drawing pedigree diagrams with R and graphviz" by Jing Hua Zhao I only want to see the highlighted nodes, but some of the applicable nodes may be buried deep within the web and not necessarily on the 'edge'.