here is a sample of my data, which is found at this link: http://www.uwyo.edu/crawford/datasets/drugreactions.txt
I made this equation for the data
fit2 <- lm(Allergens~Gender*Race*Druglevel, data=dr)
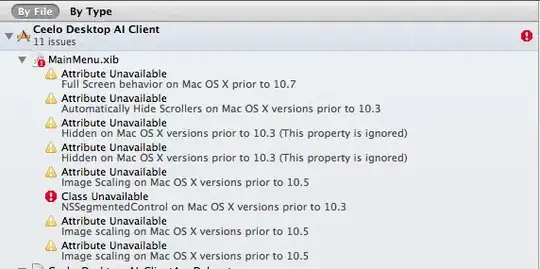
Which spit me out this
I know how to reorder the data to give a black male baseline with
dr$Race<-factor(dr$Race,levels=c("Black","Latino","Indian","Asian","NativeAmerican","Whit e"))
dr$Gender<-factor(dr$Gender,levels=c("Male","Female"))
newfit <- lm(Allergens~Gender*Race, data=dr)
However want I want is to be able to take out certain coefficients. For example, say I just want white males and black females to be in the model, instead of all the other categories. I tried
whitefit <- lm(Allergens~(Gender="Male"), data=dr)
But got an error due to uneven rows between allergens and where gender equals male.
Ideally I would like a way to take out any category so that I could completely customize the model and take out things for simplicity sake. For example, taking out male Indians from the model above.