You are using glm(...) incorrectly, which IMO is a much bigger problem than offsets.
The main underlying assumption in least squares regression is that the error in the response is normally distributed with constant variance. If the error in Y is normally distributed, then log(Y) most certainly is not. So, while you can "run the numbers" on a fit of log(Y)~X, the results will not be meaningful. The theory of generalized linear modelling was developed to deal with this problem. So using glm, rather than fit log(Y) ~X you should fit Y~X with family=poisson. The former fits
log(Y) = b0 + b1x
while the latter fits
Y = exp(b0 + b1x)
In the latter case, if the error in Y is normally distributed, and if the model is valid, then the residuals will be normally distributed, as required. Note that these two approaches give very different results for b0 and b1.
fit.incorrect <- glm(log(Y)~X,data=data2)
fit.correct <- glm(Y~X,data=data2,family=poisson)
coef(summary(fit.incorrect))
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 6.0968294 0.44450740 13.71592 0.0001636875
# X -0.2984013 0.07340798 -4.06497 0.0152860490
coef(summary(fit.correct))
# Estimate Std. Error z value Pr(>|z|)
# (Intercept) 5.8170223 0.04577816 127.06982 0.000000e+00
# X -0.2063744 0.01122240 -18.38951 1.594013e-75
In particular, the coefficient of X is almost 30% smaller when using the correct approach.
Notice how the models differ:
plot(Y~X,data2)
curve(exp(coef(fit.incorrect)[1]+x*coef(fit.incorrect)[2]),
add=T,col="red")
curve(predict(fit.correct, type="response",newdata=data.frame(X=x)),
add=T,col="blue")
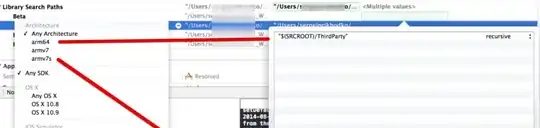
The result of the correct fit (blue curve) passes through the data more or less randomly, while the result of the incorrect fit grossly overestimates the data for small X and underestimates the data for larger X. I wonder if this is why you want to "fix" the intercept. Looking at the other answer, you can see that when you do fix Y0 = 300, the fit underestimates throughout.
In contrast, let's see what happens when we fix Y0 using glm properly.
data2$b0 <- log(300) # add the offset as a separate column
# b0 not fixed
fit <- glm(Y~X,data2,family=poisson)
plot(Y~X,data2)
curve(predict(fit,type="response",newdata=data.frame(X=x)),
add=TRUE,col="blue")
# b0 fixed so that Y0 = 300
fit.fixed <-glm(Y~X-1+offset(b0), data2,family=poisson)
curve(predict(fit.fixed,type="response",newdata=data.frame(X=x,b0=log(300))),
add=TRUE,col="green")

Here, the blue curve is the unconstrained fit (done properly), and the green curve is the fit constraining Y0 = 300. You cna see that they do not differ very much, because the correct (unconstrained) fit is already quite good.