BIG EDIT:
================
For the sake of clarity, I am removing the old results and replace it by the more recent results. The question is still the same: Am I using both Cython and Numba correctly, and what improvements to the code can be made? (I have a newer and more bare-bones temporary IPython notebook with all the code and results here)
1)
I think I figured out why there was initially no difference between Cython, Numba, and CPython: It was because I fed them
numpy arrays as input:
x = np.asarray([x_i*np.random.randint(8,12)/10 for x_i in range(n)])
instead of lists:
x = [x_i*random.randint(8,12)/10 for x_i in range(n)]
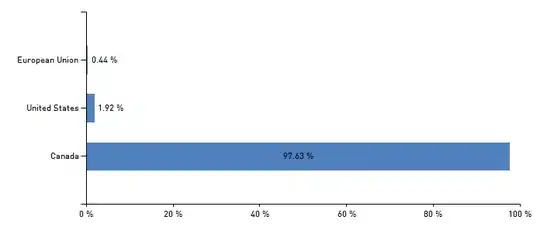
Benchmark using Numpy arrays as data input
Benchmark using Python lists as input
2)
I replaced the zip() function by explicit loops, however, it didn't make much of a difference. The code would be:
CPython
def py_lstsqr(x, y):
""" Computes the least-squares solution to a linear matrix equation. """
len_x = len(x)
x_avg = sum(x)/len_x
y_avg = sum(y)/len(y)
var_x = 0
cov_xy = 0
for i in range(len_x):
temp = (x[i] - x_avg)
var_x += temp**2
cov_xy += temp*(y[i] - y_avg)
slope = cov_xy / var_x
y_interc = y_avg - slope*x_avg
return (slope, y_interc)
Cython
%load_ext cythonmagic
%%cython
def cy_lstsqr(x, y):
""" Computes the least-squares solution to a linear matrix equation. """
cdef double x_avg, y_avg, var_x, cov_xy,\
slope, y_interc, x_i, y_i
cdef int len_x
len_x = len(x)
x_avg = sum(x)/len_x
y_avg = sum(y)/len(y)
var_x = 0
cov_xy = 0
for i in range(len_x):
temp = (x[i] - x_avg)
var_x += temp**2
cov_xy += temp*(y[i] - y_avg)
slope = cov_xy / var_x
y_interc = y_avg - slope*x_avg
return (slope, y_interc)
Numba
from numba import jit
@jit
def numba_lstsqr(x, y):
""" Computes the least-squares solution to a linear matrix equation. """
len_x = len(x)
x_avg = sum(x)/len_x
y_avg = sum(y)/len(y)
var_x = 0
cov_xy = 0
for i in range(len_x):
temp = (x[i] - x_avg)
var_x += temp**2
cov_xy += temp*(y[i] - y_avg)
slope = cov_xy / var_x
y_interc = y_avg - slope*x_avg
return (slope, y_interc)