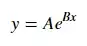I want to plot a CCDF graph for some of my simulated power-law tail data on a log-log axis, below is my R code of plotting a CCDF graph on a normal axis, I used the code on the link: (How to plot a CCDF gragh?)
> load("fakedata500.Rda")
> x<-fakedata500
> f<-ecdf(x)
> f
Empirical CDF
Call: ecdf(x)
x[1:500] = 0.50174, 0.50307, 0.50383, ..., 81.674, 140.63
> plot(f)
Below is the ECDF graph:
> plot(sort(x), 1-f(sort(x)), type="s", lwd=1)
and this command gives me the CCDF graph:

However, I would like to plot my CCDF graph on a log-log axis to produce a graph like the picture below: (graph from "Minimizing errors in identifying Lévy flight behaviour of organisms.")
Is there a way to do it in R?
If so, how to do a linear regression on the CCDF graph as well? I have tried using command below but that just not working for me.
a<-plot(sort(x), 1-f(sort(x)), type="s", lwd=1)
> a
NULL
> res=lm(a)
Error in terms.formula(formula, data = data) :
argument is not a valid model
Much appreciate.
UPDATE:
I used the code given by @BondedDust and successfully generated a CCDF graph:
(plot(sort(x) , 1-ecdf(x)(sort(x) ), log="xy"))

Below is the code of how I generated my dataset:
u<-runif(500)
fakedata500<-((2*(1-u))^(-1))