I am stuck in a rather complicated situation. I am plotting some data as an image with imshow(). Unfortunately my script is long and a little messy, so it is difficult to make a working example, but I am showing the key steps. This is how I get the data for my image from a bigger array, written in a file:
data = np.tril(np.loadtxt('IC-heatmap-20K.mtx'), 1)
#
#Here goes lot's of other stuff, where I define start and end
#
chrdata = data[start:end, start:end]
chrdata = ndimage.rotate(chrdata, 45, order=0, reshape=True,
prefilter=False, cval=0)
ax1 = host_subplot(111)
#I don't really need host_subplot() in this case, I could use something more common;
#It is just divider.append_axes("bottom", ...) is really convenient.
plt.imshow(chrdata, origin='lower', interpolation='none',
extent=[0, length*resolution, 0, length*resolution]) #resolution=20000
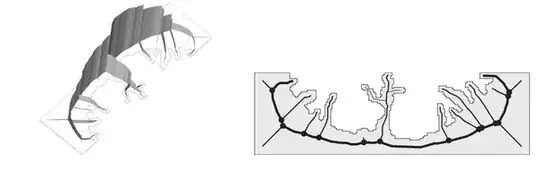
So the values I am interested in are all in a triangle with the top angle in the middle of the top side of a square. At the same time I plot some data (lot's of coloured lines in this case) along with the image near it's bottom.
So at first this looks OK, but is actually is not: all pixels in the image are not square, but elongated with their height being bigger, than their width. This is how they look if I zoom in:

This doesn't happen, If I don't set extent when calling imshow(), but I need it so that coordinates in the image and other plots (coloured lines at the bottom in this case), where identical (see Converting coordinates of a picture in matplotlib?).
I tried to fix it using aspect. I tried to do that and it fixed the pixels' shape, but I got a really weird picture:
The thing is, later in the code I explicitly set this:
ax1.set_ylim(0*resolution, length*resolution) #resolution=20000
But after setting aspect I get absolutely different y limits. And the worst thing: ax1 is now wider, than axes of another plot at the bottom, so that their coordinates do not match anymore! I add it in this way:
axPlotx = divider.append_axes("bottom", size=0.1, pad=0, sharex=ax1)
I would really appreciate help with getting it fixed: square pixels, identical coordinates in two (or more, in other cases) plots. As I see it, the axes of the image need to become wider (as aspect does), the ylims should apply and the width of the second axes should be identical to the image's. Thanks for reading this probably unclear explanation, please, let me know, if I should clarify anything.
UPDATE
As suggested in the comments, I tried to use
ax1.set(adjustable='box-forced')
And it did help with the image itself, but it caused two axes to get separated by white space. Is there any way to keep them close to each other?