I have data for a CDF in a file which looks like the following:
0.033 0.0010718113612
0.034 0.0016077170418
0.038 0.0021436227224
... ...
... ...
0.847 0.999464094319
0.862 1.0
First column is the X-axis value and the second column is the CDF value on Y-axis. I set the line style as follows:
set style line 1 lc rgb 'blue' lt 1 lw 2 pt 7 ps 0.75 # --- blue
and subsequently plot the line with the following:
plot file1 using 1:2 title 'Test Line CDF' with linespoints ls 1
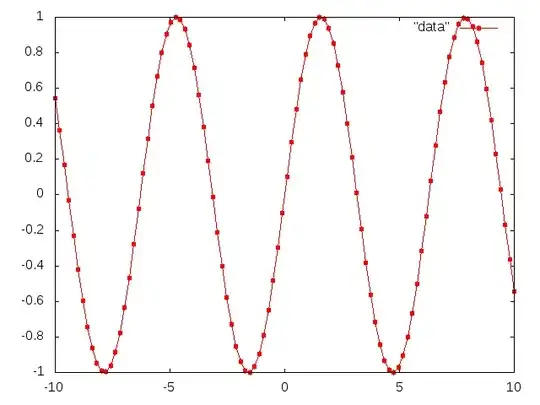
This all works fine, the problem seems to be that my CDF file is pretty big (about 250 rows) and Gnuplot would plot the marker/point (a circle in this case) for every data point. This results in a very "dense" line because of the over-concentration of markers such that the underlying line is almost not visible as I show in an example image below:

How can I selectively draw the markers so that instead of having them on all data points, I plot them after every 50 data points, without having to decrease the number of data points (which I believe is what "every n" in the plot command would do) in my data file or decrease the marker size?