data:
df <- structure(list(x = c(9.5638945103927, 13.7767187698566, 6.0019477258207,
10.1897072092089, 15.4019854273531, 10.9746646056535, 12.9429073949468,
20.7513493525379, 18.5764146937149, 2.91302077116471, 13.6523222711501,
10.0920467755108), y = c(83.949498880077, 18.066881289085, 71.3052196358606,
39.8975644317452, 57.2933166677927, 87.8484256883889, 92.6818329896141,
49.8297961197214, 56.3650103496898, 14.7950650020996, 37.9271392096266,
50.4357237591891), z = c("a", "c", "e", "f", "b", "a", "b", "a",
"b", "a", "c", "d")), .Names = c("x", "y", "z"), row.names = c(NA,
-12L), class = "data.frame")
my model:
mod <- glm(y ~ x + I(x^2) + z, family=quasipoisson, data = df)
summary(mod)
I want to plot something like this:
ggplot(df, aes(x=x,y=y)) +
geom_point() +
stat_smooth(method="lm",se=FALSE,
formula=y~x+I(x^2),fill="transparent",
colour="black") +
stat_smooth(method="lm",geom="ribbon",
formula=y~x+I(x^2),fill="transparent",
colour="red",linetype="dashed",fullrange=TRUE) +
scale_x_continuous(limits=c(-2,35)) +
coord_cartesian(xlim=c(2,25),
ylim=range(pretty(df$y)))
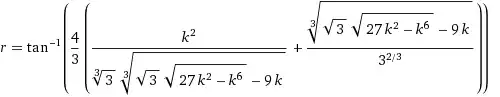
However, the model I have plotted is obviously not the same model as mod, there is no z and it is not quasiposson.
How can I plot something like the ggplot but using my actual model? I have looked at predict however, I am completely lost what to do when there is more than one explanatory, as in my case. I don't care about doing it in ggplot2