I'm working on a project using numpy and scipy and I need to fill in nanvalues. Currently I use scipy.interpolate.rbf, but it keeps causing python to crash so severely try/except won't even save it. However, after running it a few times, it seems as if it may keep failing in cases where there is data in the middle surrounded by all nans, like an island. Is there a better solution to this that won't keep crashing?
By the way, this is a LOT of data I need to extrapolate. Sometimes as much as half the image (70x70, greyscale), but it doesn't need to be perfect. It's part of an image stitching program, so as long as it's similar to the actual data, it'll work. I've tried nearest neighbor to fill in the nans, but the results are too different.
EDIT:
The image it always seems to fail on. Isolating this image allowed it to pass the image ONCE before crashing.
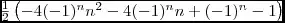
I'm using at least version NumPy 1.8.0 and SciPy 0.13.2.
