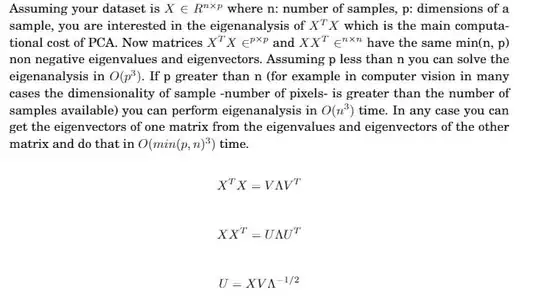
I've been reading a paper on Sparse PCA, which is: http://stats.stanford.edu/~imj/WEBLIST/AsYetUnpub/sparse.pdf
And it states that, if you have n data points, each represented with p features, then, the complexity of PCA is O(min(p^3,n^3)).
Can someone please explain how/why?