from sklearn import model_selection
test_size = 0.33
seed = 7
X_train, X_test, y_train, y_test = model_selection.train_test_split(feature_vectors, y, test_size=test_size, random_state=seed)
from sklearn.metrics import accuracy_score, f1_score, precision_score, recall_score, classification_report, confusion_matrix
model = LogisticRegression()
model.fit(X_train, y_train)
result = model.score(X_test, y_test)
print("Accuracy: %.3f%%" % (result*100.0))
y_pred = model.predict(X_test)
print("F1 Score: ", f1_score(y_test, y_pred, average="macro"))
print("Precision Score: ", precision_score(y_test, y_pred, average="macro"))
print("Recall Score: ", recall_score(y_test, y_pred, average="macro"))
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import confusion_matrix
def cm_analysis(y_true, y_pred, labels, ymap=None, figsize=(10,10)):
"""
Generate matrix plot of confusion matrix with pretty annotations.
The plot image is saved to disk.
args:
y_true: true label of the data, with shape (nsamples,)
y_pred: prediction of the data, with shape (nsamples,)
filename: filename of figure file to save
labels: string array, name the order of class labels in the confusion matrix.
use `clf.classes_` if using scikit-learn models.
with shape (nclass,).
ymap: dict: any -> string, length == nclass.
if not None, map the labels & ys to more understandable strings.
Caution: original y_true, y_pred and labels must align.
figsize: the size of the figure plotted.
"""
if ymap is not None:
# change category codes or labels to new labels
y_pred = [ymap[yi] for yi in y_pred]
y_true = [ymap[yi] for yi in y_true]
labels = [ymap[yi] for yi in labels]
# calculate a confusion matrix with the new labels
cm = confusion_matrix(y_true, y_pred, labels=labels)
# calculate row sums (for calculating % & plot annotations)
cm_sum = np.sum(cm, axis=1, keepdims=True)
# calculate proportions
cm_perc = cm / cm_sum.astype(float) * 100
# empty array for holding annotations for each cell in the heatmap
annot = np.empty_like(cm).astype(str)
# get the dimensions
nrows, ncols = cm.shape
# cycle over cells and create annotations for each cell
for i in range(nrows):
for j in range(ncols):
# get the count for the cell
c = cm[i, j]
# get the percentage for the cell
p = cm_perc[i, j]
if i == j:
s = cm_sum[i]
# convert the proportion, count, and row sum to a string with pretty formatting
annot[i, j] = '%.1f%%\n%d/%d' % (p, c, s)
elif c == 0:
annot[i, j] = ''
else:
annot[i, j] = '%.1f%%\n%d' % (p, c)
# convert the array to a dataframe. To plot by proportion instead of number, use cm_perc in the DataFrame instead of cm
cm = pd.DataFrame(cm, index=labels, columns=labels)
cm.index.name = 'Actual'
cm.columns.name = 'Predicted'
# create empty figure with a specified size
fig, ax = plt.subplots(figsize=figsize)
# plot the data using the Pandas dataframe. To change the color map, add cmap=..., e.g. cmap = 'rocket_r'
sns.heatmap(cm, annot=annot, fmt='', ax=ax)
#plt.savefig(filename)
plt.show()
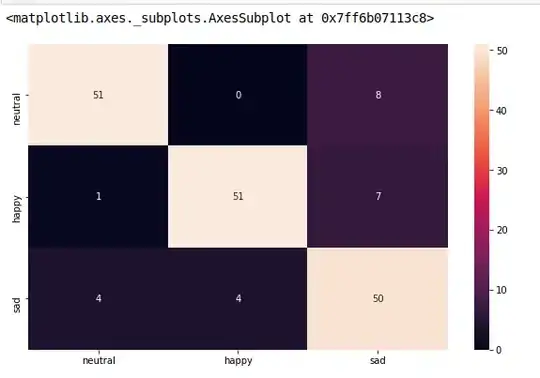
cm_analysis(y_test, y_pred, model.classes_, ymap=None, figsize=(10,10))
using https://gist.github.com/hitvoice/36cf44689065ca9b927431546381a3f7
Note that if you use rocket_r it will reverse the colors and somehow it looks more natural and better such as below: