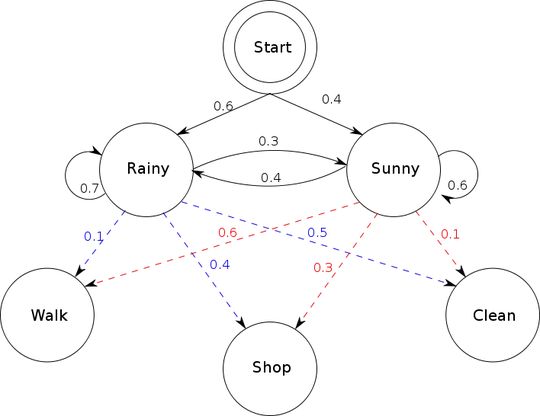
I think the question is clear enough. I want to make a hidden Markov model in Python and draw a vizualization model of it. So, it's something like this picture:
Is there any module to do that? I've googled it and found nothing.
I think the question is clear enough. I want to make a hidden Markov model in Python and draw a vizualization model of it. So, it's something like this picture:

Is there any module to do that? I've googled it and found nothing.
The dot package from graphviz is the best I've found. The syntax is simple, simpler than xml.
Though I've never worked with Hidden Markov Models, when I need to visualize a graph (directed, with labels, colors, etc.), I use Gephi, a GUI graph browser/editor and generate the graphs programmatically as GraphML files, which is an XML-based format. Python has good XML-handling tools (in the standard library and lxml). Gephi recognizes some of the <data> sub-elements as positions, colors, and labels for nodes and edges.
The Python library pomegranate has good support for Hidden Markov Models. It includes functionality for defining such models, learning it from data, doing inference, and visualizing the transitions graph (as you request here).
Below is example code for defining a model, and plotting the states and transitions. The image output will be like this:
from pomegranate import HiddenMarkovModel, State, DiscreteDistribution
from matplotlib import pyplot as plt
def build_model():
d1 = DiscreteDistribution({'A' : 0.50, 'B' : 0.50})
d2 = DiscreteDistribution({'A' : 0.10, 'B' : 0.90})
d3 = DiscreteDistribution({'A' : 0.90, 'B' : 0.10})
s1 = State(d1, name="s1")
s2 = State(d2, name="s2")
s3 = State(d3, name="s3")
model = HiddenMarkovModel(name='my model')
model.add_states(s1, s2, s3)
model.add_transition(model.start, s1, 1.0)
model.add_transition(s1, s1, 0.7)
model.add_transition(s1, s2, 0.3) # s1->s2
model.add_transition(s2, s2, 0.8)
model.add_transition(s2, s3, 0.0) # no transition from s2 to s3
model.add_transition(s1, s3, 0.1) # indirect from s1 to s3
model.add_transition(s3, s1, 0.1) # indirect from s3 to s1
model.add_transition(s3, s3, 0.9)
model.add_transition(s3, model.end, 0.1)
model.start.name = 'start'
model.end.name = 'end'
model.bake()
return model
model = build_model()
fig, ax = plt.subplots(1)
model.plot(ax=ax, precision=2)
fig.savefig('model.png')