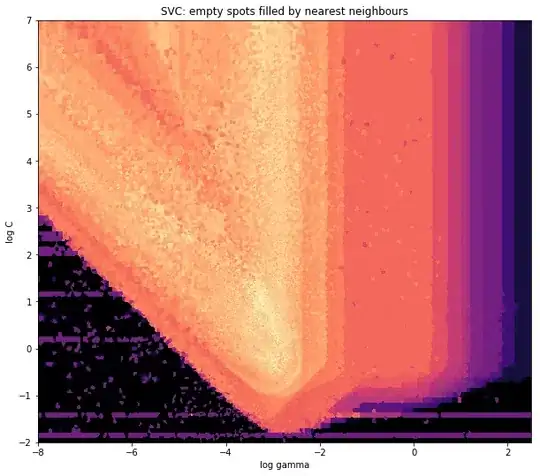
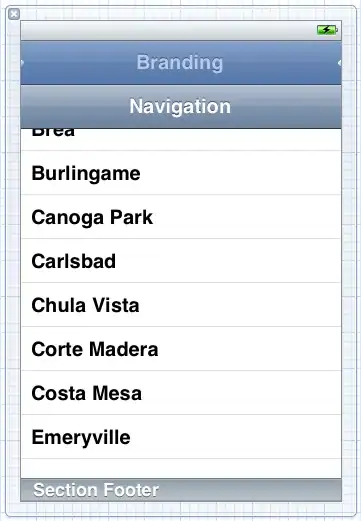
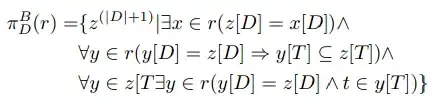but you need to set the vertex positions pretty much by hand.
This is not totally correct. I will show an easier interactive way. I will use the below code by Greg Snow.
dynmodfunc <- function() {
plot(0:1,0:1,ann=FALSE,type='n')
mypoints <- matrix(ncol=2, nrow=0)
while( length(p <- locator(1, type='p', col='red')) ) {
mypoints <- rbind(mypoints, unlist(p))
plot(mypoints, col='red', ann=FALSE, xlim=0:1, ylim=0:1)
if(nrow(mypoints)>1) {
xspline(mypoints, shape=-1)
}
}
mypoints
}
(out <- dynmodfunc())
I want to plot this dominance graph
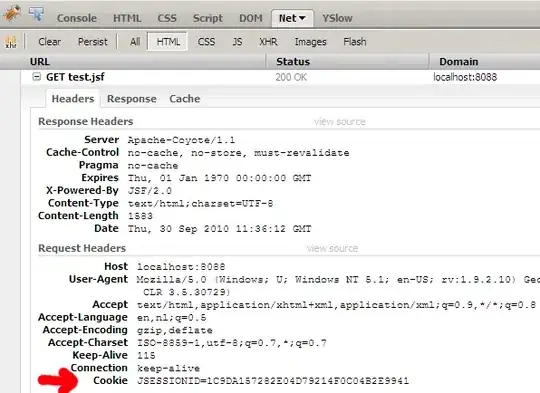
but adjm<-t(matrix(c(0,0,0,0,1,1,0, 0,0,0,0,1,1,0, 0,0,0,0,1,1,0, 0,0,0,0,1,1,0, 0,0,0,0,0,0,0, 0,0,0,0,0,0,0, 1,1,1,1,0,0,0),nrow=7,ncol=7)); g1<-graph.adjacency(adjm); plot(g1) will generate
and use Greg's code: click points on the GUI and then click right-mouse-button
where I have the points in the order of 7-1-2-3-4-5-6 as shown by the line or by the debug:
> out
x y
[1,] 0.5082585 1.03551763
[2,] 0.1067841 0.59191675
[3,] 0.3818711 0.59358184
[4,] 0.6380311 0.58883584
[5,] 0.8787300 0.58464820
[6,] 0.3417308 0.09010765
[7,] 0.6614686 0.07504212
> str(out);dput(out)
num [1:7, 1:2] 0.508 0.107 0.382 0.638 0.879 ...
- attr(*, "dimnames")=List of 2
..$ : NULL
..$ : chr [1:2] "x" "y"
structure(c(0.508258492696219, 0.106784127536735, 0.381871061286441,
0.63803114223351, 0.878729976271015, 0.341730770349654, 0.661468641881514,
1.03551763062279, 0.591916752784156, 0.593581838904923, 0.588835844931958,
0.584648203191106, 0.0901076547476853, 0.0750421150561933), .Dim = c(7L,
2L), .Dimnames = list(NULL, c("x", "y")))
and now by Gabor Csardi's solution it should be possible to get the levelled plot. We have now the co-occurrency matrix ajdm and coordinates coords for each point. I cannot yet see how to use them with Gabor's method, trying to understand.
Code for fast copy-pasting
> library(igraph);
> coords<-structure(c(0.508258492696219, 0.106784127536735, 0.381871061286441, 0.63803114223351, 0.878729976271015, 0.341730770349654, 0.661468641881514, 1.03551763062279, 0.591916752784156, 0.593581838904923, 0.588835844931958, 0.584648203191106, 0.0901076547476853, 0.0750421150561933), .Dim = c(7L, 2L), .Dimnames = list(NULL, c("x", "y")))
> adjm<-t(matrix(c(0,0,0,0,1,1,0, 0,0,0,0,1,1,0, 0,0,0,0,1,1,0, 0,0,0,0,1,1,0, 0,0,0,0,0,0,0, 0,0,0,0,0,0,0, 1,1,1,1,0,0,0),nrow=7,ncol=7)); g1<-graph.adjacency(adjm); plot(g1)