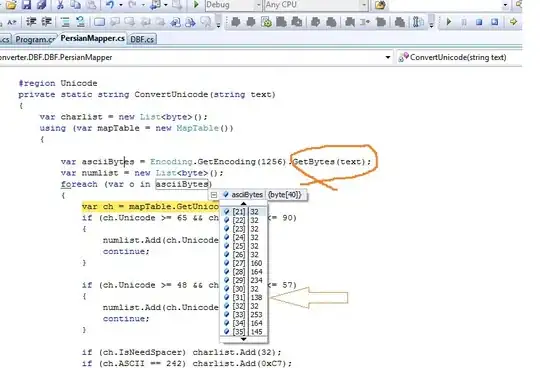
The parallel computation of the LCS follows the wavefront pattern. Here is the parallel function that is slower than a serial implementation. (I think that the number of diagonals(parallel) vs number of rows(serial) has something to do with it)
void parallelLCS(char * sequence_a, char * sequence_b, size_t size_a, size_t size_b) {
double start, end;
int ** dp_table = new int*[size_a + 1];
for (int i = 0; i <= size_a; i++)
dp_table[i] = new int[size_b + 1];
for (int i = 1; i <= size_a; i++)
dp_table[i][0] = 0;
for (int j = 0; j <= size_b; j++)
dp_table[0][j] = 0;
int p_threads = 2;
int diagonals = size_a + size_b;
start = omp_get_wtime();
#pragma omp parallel num_threads(p_threads) default(none) firstprivate(p_threads,size_a,size_b,sequence_a,sequence_b) shared(dp_table,diagonals)
{
for (int curr_diagonal = 1; curr_diagonal <= (diagonals - 1);) {
int j = omp_get_thread_num() + 1; //column index
int i = curr_diagonal - j + 1; //row index
for (; j <= curr_diagonal; j += p_threads, i = i - p_threads) {
if (i <= size_a && j <= size_b) {
if (sequence_a[i] == sequence_b[j]) {
dp_table[i][j] = dp_table[i - 1][j - 1] + 1;
} else if (dp_table[i - 1][j] >= dp_table[i][j - 1]) {
dp_table[i][j] = dp_table[i - 1][j];
} else {
dp_table[i][j] = dp_table[i][j - 1];
}
}
}
curr_diagonal++;
#pragma omp barrier
}
}
end = omp_get_wtime();
printf("\nParallel - Final answer: %d\n", dp_table[size_a][size_b]);
printf("Time: %f\n", end - start);
//Delete dp_table
for (int i = 0; i <= size_a; i++)
delete [] dp_table[i];
delete [] dp_table;
}
and here is the serial function
void serialLCS(char * sequence_a, char * sequence_b, size_t size_a, size_t size_b) {
double start, end;
int ** dp_table = new int*[size_a + 1];
for (int i = 0; i <= size_a; i++)
dp_table[i] = new int[size_b + 1];
for (int i = 1; i <= size_a; i++)
dp_table[i][0] = 0;
for (int j = 0; j <= size_b; j++)
dp_table[0][j] = 0;
start = omp_get_wtime();
for (int i = 1; i <= size_a; i++) {
for (int j = 1; j <= size_b; j++) {
if (sequence_a[i] == sequence_b[j]) {
dp_table[i][j] = dp_table[i - 1][j - 1] + 1;
} else if (dp_table[i - 1][j] >= dp_table[i][j - 1]) {
dp_table[i][j] = dp_table[i - 1][j];
} else {
dp_table[i][j] = dp_table[i][j - 1];
}
}
}
end = omp_get_wtime();
printf("\nSerial - Final answer: %d\n", dp_table[size_a][size_b]);
printf("Time: %f\n", end - start);
//Delete dp_table
for (int i = 0; i <= size_a; i++)
delete [] dp_table[i];
delete [] dp_table;
}
...thought I'd add the test function
#include <cstdlib>
#include <stdio.h>
#include <omp.h>
void serialLCS(char * sequence_a, char * sequence_b, size_t size_a, size_t size_b);
void parallelLCS(char * sequence_a, char * sequence_b, size_t size_a, size_t size_b);
int main() {
size_t size_a;
size_t size_b;
printf("Enter size of sequence A: ");
scanf("%zd",&size_a);
printf("Enter size of sequence B: ");
scanf("%zd",&size_b);
//keep larger sequence in sequence_a
if (size_b > size_a) size_a ^= size_b ^= size_a ^= size_b;
char * sequence_a = new char[size_a + 1];
char * sequence_b = new char[size_b + 1];
sequence_a[0] = sequence_b[0] = '0';
const size_t alphabet_size = 12;
char A[alphabet_size] = {'A', 'T', 'G', 'C', 'Q', 'W', 'E', 'R', 'Y', 'U', 'I', 'O'};
char AA[alphabet_size] = {'T', 'C', 'A', 'G', 'R', 'E', 'W', 'Q', 'O', 'I', 'U', 'Y'};
for (size_t i = 1; i < size_a; i++) {
sequence_a[i] = A[rand() % alphabet_size];
}
for (size_t i = 1; i < size_b; i++) {
sequence_b[i] = AA[rand() % alphabet_size];
}
serialLCS(sequence_a, sequence_b, size_a, size_b);
parallelLCS(sequence_a, sequence_b, size_a, size_b);
delete [] sequence_a;
delete [] sequence_b;
return 0;
}