how do I select a subset of points at a regular density? More formally,
Given
- a set A of irregularly spaced points,
- a metric of distance
dist(e.g., Euclidean distance), - and a target density d,
how can I select a smallest subset B that satisfies below?
- for every point x in A,
- there exists a point y in B
- which satisfies
dist(x,y) <= d
My current best shot is to
- start with A itself
- pick out the closest (or just particularly close) couple of points
- randomly exclude one of them
- repeat as long as the condition holds
and repeat the whole procedure for best luck. But are there better ways?
I'm trying to do this with 280,000 18-D points, but my question is in general strategy. So I also wish to know how to do it with 2-D points. And I don't really need a guarantee of a smallest subset. Any useful method is welcome. Thank you.
bottom-up method
- select a random point
- select among unselected
yfor whichmin(d(x,y) for x in selected)is largest - keep going!
I'll call it bottom-up and the one I originally posted top-down. This is much faster in the beginning, so for sparse sampling this should be better?
performance measure
If guarantee of optimality is not required, I think these two indicators could be useful:
- radius of coverage:
max {y in unselected} min(d(x,y) for x in selected) - radius of economy:
min {y in selected != x} min(d(x,y) for x in selected)
RC is minimum allowed d, and there is no absolute inequality between these two. But RC <= RE is more desirable.
my little methods
For a little demonstration of that "performance measure," I generated 256 2-D points distributed uniformly or by standard normal distribution. Then I tried my top-down and bottom-up methods with them. And this is what I got:
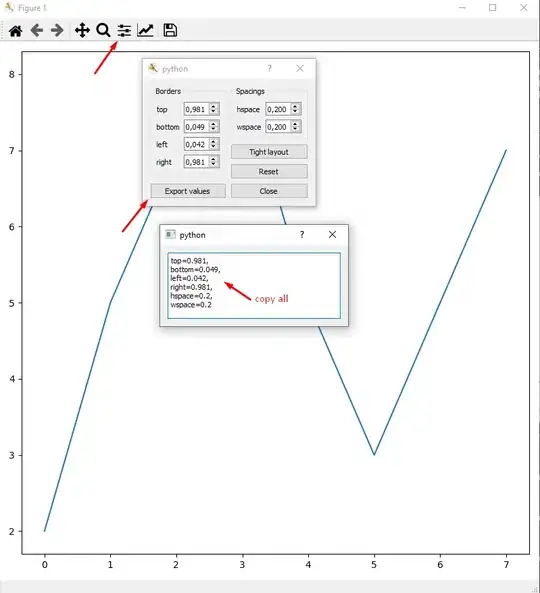
RC is red, RE is blue. X axis is number of selected points. Did you think bottom-up could be as good? I thought so watching the animation, but it seems top-down is significantly better (look at the sparse region). Nevertheless, not too horrible given that it's much faster.
Here I packed everything.
http://www.filehosting.org/file/details/352267/density_sampling.tar.gz