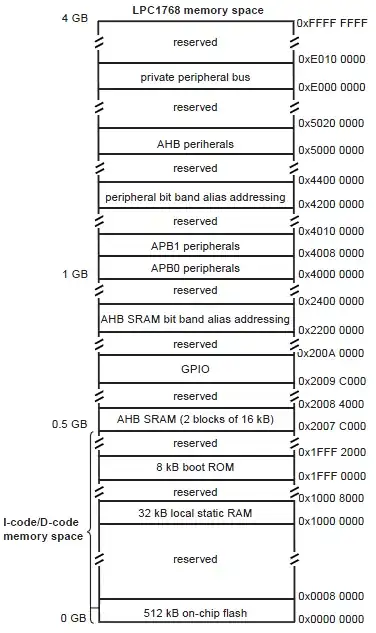
This is how I would do it:
- Create a kernel, it defines a pixels neighbourhood.
- Create a new image by dilating your image using this kernel. This dilated image contains the maximum neighbourhood value for every point.
- Do an equality comparison between these two arrays. Wherever they are equal is a valid neighbourhood maximum, and is set to
255 in the comparison array.
- Multiply the comparison array, and the original array together (scaling appropriately).
- This is your final array, containing only neighbourhood maxima.
This is illustrated by these zoomed in images:
9 pixel by 9 pixel original image:

After processing with a 5 by 5 pixel kernel, only the local neighbourhood maxima remain (ie. maxima seperated by more than 2 pixels from a pixel with a greater value):
There is one caveat. If two nearby maxima have the same value then they will both be present in the final image.
Here is some Python code that does it, it should be very easy to convert to c++:
import cv
im = cv.LoadImage('fish2.png',cv.CV_LOAD_IMAGE_GRAYSCALE)
maxed = cv.CreateImage((im.width, im.height), cv.IPL_DEPTH_8U, 1)
comp = cv.CreateImage((im.width, im.height), cv.IPL_DEPTH_8U, 1)
#Create a 5*5 kernel anchored at 2,2
kernel = cv.CreateStructuringElementEx(5, 5, 2, 2, cv.CV_SHAPE_RECT)
cv.Dilate(im, maxed, element=kernel, iterations=1)
cv.Cmp(im, maxed, comp, cv.CV_CMP_EQ)
cv.Mul(im, comp, im, 1/255.0)
cv.ShowImage("local max only", im)
cv.WaitKey(0)
I didn't realise until now, but this is what @sansuiso suggested in his/her answer.
This is possibly better illustrated with this image, before:

after processing with a 5 by 5 kernel:
solid regions are due to the shared local maxima values.



