I am using Metpy's interpolate_to_grid function to plot contours for US upper air station data. I have been getting odd results using the "cressman" and "barnes" interpolation methods, as will be seen below. I believe these methods should give better results for upper-air station data than I am getting, so I would like to know how best to use the "cressman" and "barnes" interpolation methods for this kind of problem: upper air station data, rather than model data. Thank you!
NOTE: Updated 12/11/2022 Question Partially Answered (see below)
The Problem
Consider the following inputs, with (x,y) representing station latitudes and longitudes converted using Cartopy's LambertConformal transformation, and instead of actual station data, we use z, representing a better-behaved paraboloidal function.
x = np.array([-588983.27466878, -597509.66039475, -593548.25150405,
-708631.9755505 , 141572.74682356, -255553.18180321,
2605.75272771, -151524.57820357, -203291.23499391,
-87463.57692061, -22995.70471404, 374092.71512281,
214697.3152085 , 169183.52087143, 700245.60311235,
297722.11595698, 258836.14441127, 463635.45150329,
495809.6650171 , 941489.32668957, 1187111.28831832,
945411.2861692 , 861812.39100534, 535475.69810851,
508126.79898721, 647449.62111219, 919682.88858901,
1017240.1399329 , 1172517.98849875])
y = np.array([ -680251.55108761, -330509.2160825 , 24118.72712136,
888634.46530843, 1031366.82096134, 1000957.63129123,
302813.43368973, 25326.16666977, -331119.27031642,
-616634.39369923, -1001199.94104898, 452311.04341359,
46298.26388061, -302270.45196599, -3201.83555954,
-668399.94426101, -994537.91752629, 1173948.32332572,
1513810.0688783 , 1096427.60089224, 217056.91961851,
451671.20085842, 665507.85533899, 221907.02519804,
-494811.23266806, -512631.01355181, -248034.64814128,
-566019.80063491, -154148.98796047])
# synthetic data describing paraboloid, scaled by 1e-11 to get z into the range [0,25]
z = np.zeros(len(x))
xmean = np.mean(x)
ymean = np.mean(y)
z = 1e-11*((x-xmean)**2 + (y-ymean)**2)
# This should be symmetric about (roughly) the center of the region of interest
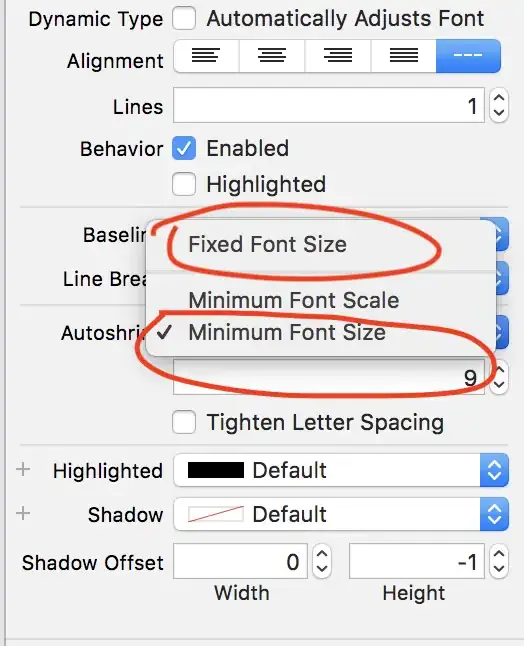
Now interpolate that data to a grid using the "barnes" method, allowing the function to select the default search_radius. Also plot the station locations in red.
# Interpolate z to a regular grid using MetPy's interpolate_to_grid
X,Y,Z = interpolate_to_grid(x,y,z,interp_type='barnes',hres=25000)
Levels = np.arange(np.floor(np.nanmin(Z)),np.ceil(np.nanmax(Z)))
fig0 = plt.figure(figsize=(7.5,7.5))
fig0.patch.set_facecolor('white')
fig0.patch.set_alpha(1)
ax0 = fig0.add_subplot(1,1,1)
plt.contour(X,Y,Z,levels=Levels,colors='gray',linewidths=.5)
cf0 = ax0.contour(X,Y,Z,alpha=.75,levels=Levels,colors='gray',linewidths=.5)
ax0.clabel(cf0, cf0.levels, inline = True, fontsize=10, colors='black', fmt='%.0f')
plt.scatter(x,y,s=5,color = 'red')
fig0.savefig('Fig1.png',bbox_inches='tight',dpi=(144))
plt.show()
## OUTPUT ##
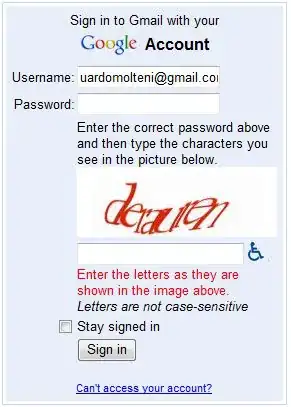
Clearly this does not represent a paraboloid, although the contours do suggest a minimum in a concave-up surface near the center of the region of interest. To make sure we are doing things reasonably, try a simpler interpolation function (cubic).
# Try a simpler interpolation function
X,Y,Z1 = interpolate_to_grid(x,y,z,interp_type='cubic',hres=25000)
Level1s = np.arange(np.floor(np.nanmin(Z1)),np.ceil(np.nanmax(Z1)))
fig0 = plt.figure(figsize=(7.5,7.5))
fig0.patch.set_facecolor('white')
fig0.patch.set_alpha(1)
ax0 = fig0.add_subplot(1,1,1)
cf0 = ax0.contour(X,Y,Z1,alpha=.75,levels=Level1s,colors='gray',linewidths=.5)
ax0.clabel(cf0, cf0.levels, inline = True, fontsize=10, colors='black', fmt='%.0f')
plt.scatter(x,y,s=5,color = 'red')
fig0.savefig('Fig2.png',bbox_inches='tight',dpi=(144))
plt.show()
## OUTPUT ##
This is a lot more like what we expected to see (ignoring the incomplete contours near the edges due to nan's in the interpolated data, which are not an issue at this point.)
We get a similar result using the "linear" method (not shown), although predictably the contours are not as smooth.
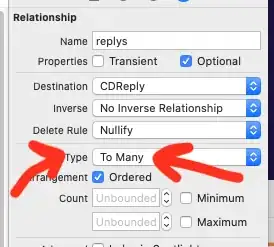
Trying the "cressman" method, we do get a more reasonable set of contours, but still not as good as found using the cubic and linear methods:
# Try the cressman interpolation function
X,Y,Z3 = interpolate_to_grid(x,y,z,interp_type='cressman',hres=25000)
Level3s = np.arange(np.floor(np.nanmin(Z3)),np.ceil(np.nanmax(Z3)))
fig0 = plt.figure(figsize=(7.5,7.5))
fig0.patch.set_facecolor('white')
fig0.patch.set_alpha(1)
ax0 = fig0.add_subplot(1,1,1)
cf0 = ax0.contour(X,Y,Z3,alpha=.75,levels=Level3s,colors='gray',linewidths=.5)
ax0.clabel(cf0, cf0.levels, inline = True, fontsize=10, colors='black', fmt='%.0f')
plt.scatter(x,y,s=5,color = 'red')
fig0.savefig('Fig3.png',bbox_inches='tight',dpi=(144))
plt.show()
## OUTPUT ##
The cressman method does provide a better representation of the test paraboloid, but the overall scale is wrong: the minimum of the cressman-based surface is ~5 (compared to 0 for the test surface), and a maximum ~13 (compared to ~20 for the test surface.) Also, the cressman-minimum is too broad and not centered on the region of interest.
Finally: adding search_radius as a parameter (for the cressman and barnes methods), and increasing the search_radius stepwise to 2000000 did not resolve the issues we see above. Note, however, that values above 1250000 led to increasingly bad performance. (For brevity, these results are not shown.)
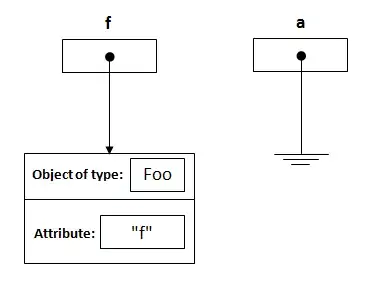
Update 12/11/2022. Question Partially Answered
After experimenting with the additional barnes-related input parameters gamma and kappa_star, we are getting more reasonable results.
# Interpolate z to a regular grid using MetPy's interpolate_to_grid, adding gamma and kappa_star parameters:
X,Y,Z = interpolate_to_grid(x,y,z,interp_type='barnes',hres=100000,gamma=5,kappa_star=1)
Levels = np.arange(np.floor(np.nanmin(Z)),np.ceil(np.nanmax(Z)))
fig0 = plt.figure(figsize=(7.5,7.5))
fig0.patch.set_facecolor('white')
fig0.patch.set_alpha(1)
ax0 = fig0.add_subplot(1,1,1)
plt.contour(X,Y,Z,levels=Levels,colors='gray',linewidths=.5)
cf0 = ax0.contour(X,Y,Z,alpha=.75,levels=Levels,colors='gray',linewidths=.5)
ax0.clabel(cf0, cf0.levels, inline = True, fontsize=8, colors='gray', fmt='%.0f')
plt.scatter(x,y,s=5,color = 'red')
for i in np.arange(len(z)):
plt.text(x[i],y[i],np.round(z[i],1),fontsize=10)
fig0.savefig('Fig4.png',bbox_inches='tight',dpi=(144))
plt.show()