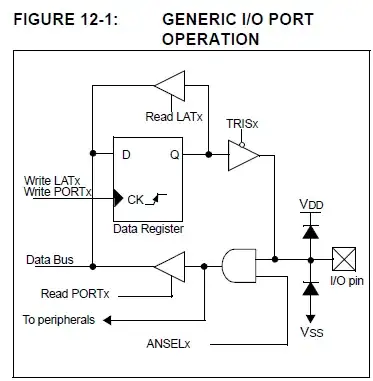
I have a Panda Dataframe df in input to Pycaret library. So the df has :
3 categoricals variables:
LIB_SOURCE : values: 'arome_001', 'gfs_025' and 'arpege_01'
MonthNumber : values from 1 to 12
origine : 'Sencrop' and 'Visiogreen' values
3 continuous variables :
TEMPERATURE_PREDITE DIFF_HOURS TEMPERATURE_OBSERVEE
I let Pycaret encoding categorical features to 0/1 and manage multicollinearity:
regression = setup(data = dataset_predictions_meteo,
target = 'TEMPERATURE_PREDITE',
categorical_features = ['MonthNumber' , 'origine' , 'LIB_SOURCE'],
numeric_features = ['DIFF_HOURS' , 'TEMPERATURE_OBSERVEE'],
session_id=123,
train_size=0.8,
normalize=True,
#transform_target=True,
remove_perfect_collinearity = True
)
But as you can see in the screen above, Pycaret doesn't well manage multicollinearity : PyCaret should remove by itself 1 of 3 columns 'arome_001', 'gfs_025' and 'arpege_01' (get_config('X')). But PyCaret keeps all 3 columns.
Why PyCaret doesn't remove one of 3 columns? Thanks.