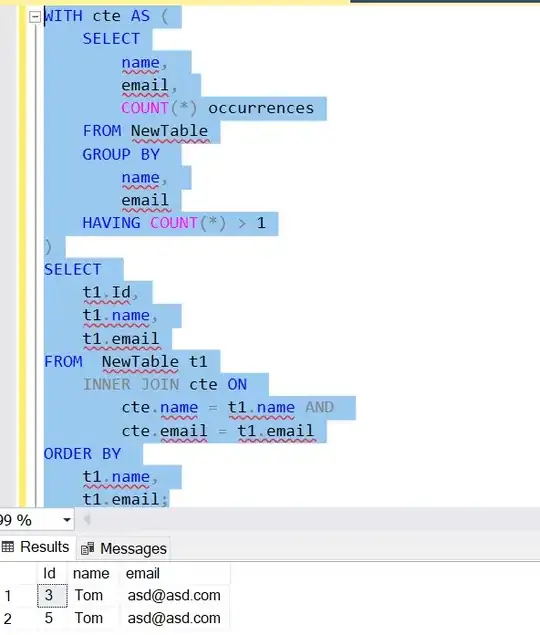
I have a set of points on a map, each with a given parameter value. I would like to:
- Cluster them spatially and ignore any clusters having fewer than 10 points. My df should have a column (Clust) for the cluster each point belongs to [DONE]
- Sub-cluster the parameter values within each cluster; add a column to my df (subClust) used to categorize each point by sub-cluster.
I don't know how to do the second part, except maybe with loops.
The image shows the set of spatially distributed points (top left) colour coded by cluster and sorted by parameter value in the top right plot. The bottom row shows clusters with >10 points (left) and facets for each cluster sorted by parameter value (right). It's these facets that I'd like to be able to colour code by sub-cluster according to a minimum cluster separation distance (d=1)
Any pointers/help appreciated. My reproducible code is below.
# TESTING
library(tidyverse)
library(gridExtra)
# Create a random (X, Y, Value) dataset
set.seed(36)
x_ex <- round(rnorm(200,50,20))
y_ex <- round(runif(200,0,85))
values <- rexp(200, 0.2)
df_ex <- data.frame(ID=1:length(y_ex),x=x_ex,y=y_ex,Test_Param=values)
# Cluster data by (X,Y) location
d = 4
chc <- hclust(dist(df_ex[,2:3]), method="single")
# Distance with a d threshold - used d=40 at one time but that changes...
chc.d40 <- cutree(chc, h=d)
# max(chc.d40)
# Join results
xy_df <- data.frame(df_ex, Clust=chc.d40)
# Plot results
breaks = max(chc.d40)
xy_df_filt <- xy_df %>% dplyr::group_by(Clust) %>% dplyr::mutate(n=n()) %>% dplyr::filter(n>10)# %>% nrow
p1 <- ggplot() +
geom_point(data=xy_df, aes(x=x, y=y, colour = Clust)) +
scale_color_gradientn(colours = rainbow(breaks)) +
xlim(0,100) + ylim(0,100)
p2 <- xy_df %>% dplyr::arrange(Test_Param) %>%
ggplot() +
geom_point(aes(x=1:length(Test_Param),y=Test_Param, colour = Test_Param)) +
scale_colour_gradient(low="red", high="green")
p3 <- ggplot() +
geom_point(data=xy_df_filt, aes(x=x, y=y, colour = Clust)) +
scale_color_gradientn(colours = rainbow(breaks)) +
xlim(0,100) + ylim(0,100)
p4 <- xy_df_filt %>% dplyr::arrange(Test_Param) %>%
ggplot() +
geom_point(aes(x=1:length(Test_Param),y=Test_Param, colour = Test_Param)) +
scale_colour_gradient(low="red", high="green") +
facet_wrap(~Clust, scales="free")
grid.arrange(p1, p2, p3, p4, ncol=2, nrow=2)
THIS SNIPPET DOES NOT WORK - can't pipe within dplyr mutate() ...
# Second Hierarchical Clustering: Try to sub-cluster by Test_Param within the individual clusters I've already defined above
xy_df_filt %>% # This part does not work
dplyr::group_by(Clust) %>%
dplyr::mutate(subClust = hclust(dist(.$Test_Param), method="single") %>%
cutree(, h=1))
Below is a way around it using a loop - but I'd really rather learn how to do this using dplyr or some other non-loop method. An updated image showing the sub-clustered facets follows.
sub_df <- data.frame()
for (i in unique(xy_df_filt$Clust)) {
temp_df <- xy_df_filt %>% dplyr::filter(Clust == i)
# Cluster data by (X,Y) location
a_d = 1
a_chc <- hclust(dist(temp_df$Test_Param), method="single")
# Distance with a d threshold - used d=40 at one time but that changes...
a_chc.d40 <- cutree(a_chc, h=a_d)
# max(chc.d40)
# Join results to main df
sub_df <- bind_rows(sub_df, data.frame(temp_df, subClust=a_chc.d40)) %>% dplyr::select(ID, subClust)
}
xy_df_filt_2 <- left_join(xy_df_filt,sub_df, by=c("ID"="ID"))
p4 <- xy_df_filt_2 %>% dplyr::arrange(Test_Param) %>%
ggplot() +
geom_point(aes(x=1:length(Test_Param),y=Test_Param, colour = subClust)) +
scale_colour_gradient(low="red", high="green") +
facet_wrap(~Clust, scales="free")
grid.arrange(p1, p2, p3, p4, ncol=2, nrow=2)