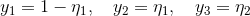What you are describing is generally referred to as "variable aggregation." As you indicate, there are four basic steps:
- Identify the linear equality equations you want to remove
- Compute the substitution map
- Deactivate the equality constraints that you want to remove
- Substitute variables on all remaining constraints
It sounds like you have 1 and 2 under control. For 3, assuming you identified a Constraint m.c you want to deactivate, you just need to call m.c.deactivate().
For 4, you will want to generate new expressions for the remaining Constraint "body" expressions (variables only appear in the body and not in the lower/upper bounds). For current Pyomo releases (through 5.4.x), you can perform variable substitution by leveraging the clone_expression(). You need to generate a "substitution map": a dict that maps the id() of the variables you want to the new expression you want to use. For example:
from pyomo.core.base.expr import clone_expression
m = ConcreteModel()
m.y = Var([1,2,3])
m.eta = Var([1,2])
# ...
m.c = Constraint(expr=m.y[1]**2 + m.y[3]**2 <= 4)
# ...
substitution_map = {
id(m.y[1]): 1 - m.eta[1],
id(m.y[2]): m.eta[1],
id(m.y[3]): m.eta[2],
}
m.c = (m.c.lower, clone_expression(m.c.body, substitute=substitution_map), m.c.upper)
Finally, the disclaimers:
- Setting the constraint with this syntax should work with recent Pyomo releases (I tested back through 5.1)
- This approach technically violates one of the assumptions in the current Pyomo expression system (it generates potentially "entangled" expressions: expressions that share common sub-trees). While not "good", it shouldn't cause troubles, unless you do additional transformations / expression manipulation.
- Pyomo 5.5 will have a new expression system that will likely have a different mechanism for manipulating / substituting variables.